A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes - PubMed (original) (raw)
A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes
R Alexander Pyron et al. BMC Evol Biol. 2013.
Abstract
Background: The extant squamates (>9400 known species of lizards and snakes) are one of the most diverse and conspicuous radiations of terrestrial vertebrates, but no studies have attempted to reconstruct a phylogeny for the group with large-scale taxon sampling. Such an estimate is invaluable for comparative evolutionary studies, and to address their classification. Here, we present the first large-scale phylogenetic estimate for Squamata.
Results: The estimated phylogeny contains 4161 species, representing all currently recognized families and subfamilies. The analysis is based on up to 12896 base pairs of sequence data per species (average = 2497 bp) from 12 genes, including seven nuclear loci (BDNF, c-mos, NT3, PDC, R35, RAG-1, and RAG-2), and five mitochondrial genes (12S, 16S, cytochrome b, ND2, and ND4). The tree provides important confirmation for recent estimates of higher-level squamate phylogeny based on molecular data (but with more limited taxon sampling), estimates that are very different from previous morphology-based hypotheses. The tree also includes many relationships that differ from previous molecular estimates and many that differ from traditional taxonomy.
Conclusions: We present a new large-scale phylogeny of squamate reptiles that should be a valuable resource for future comparative studies. We also present a revised classification of squamates at the family and subfamily level to bring the taxonomy more in line with the new phylogenetic hypothesis. This classification includes new, resurrected, and modified subfamilies within gymnophthalmid and scincid lizards, and boid, colubrid, and lamprophiid snakes.
Figures
Figure 1
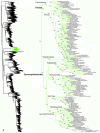
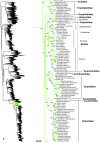
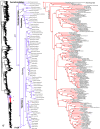
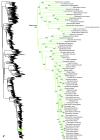
Higher-level squamate phylogeny. Skeletal representation of the 4161-species tree from maximum-likelihood analysis of 12 genes, with tips representing families and subfamilies (following our taxonomic revision; species considered incertae sedis are not shown). Numbers at nodes are SHL values greater than 50%. The full tree is presented in Figures 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28.
Figure 2
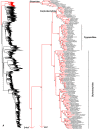
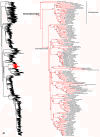
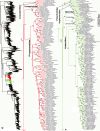
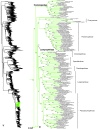
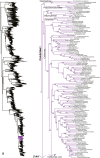
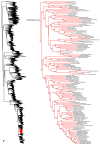
Species-level squamate phylogeny. Large-scale maximum likelihood estimate of squamate phylogeny, containing 4161 species. Numbers at nodes are SHL values greater than 50%. A skeletal version of this tree is presented in Figure 1. Bold italic letters indicate figure panels (A-AA). Within panels, branch lengths are proportional to expected substitutions per site, but the relative scale differs between panels.
Figure 3
Species-level squamate phylogeny continued (B).
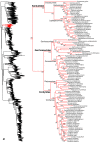
Figure 4
Species-level squamate phylogeny continued (C).
Figure 5
Species-level squamate phylogeny continued (D).
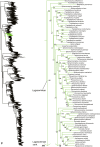
Figure 6
Species-level squamate phylogeny continued (E).
Figure 7
Species-level squamate phylogeny continued (F).
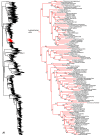
Figure 8
Species-level squamate phylogeny continued (G).
Figure 9
Species-level squamate phylogeny continued (H).
Figure 10
Species-level squamate phylogeny continued (I).
Figure 11
Species-level squamate phylogeny continued (J).
Figure 12
Species-level squamate phylogeny continued (K).
Figure 13
Species-level squamate phylogeny continued (L).
Figure 14
Species-level squamate phylogeny continued (M).
Figure 15
Species-level squamate phylogeny continued (N).
Figure 16
Species-level squamate phylogeny continued (O).
Figure 17
Species-level squamate phylogeny continued (P).
Figure 18
Species-level squamate phylogeny continued (Q).
Figure 19
Species-level squamate phylogeny continued (R).
Figure 20
Species-level squamate phylogeny continued (S).
Figure 21
Species-level squamate phylogeny continued (T).
Figure 22
Species-level squamate phylogeny continued (U).
Figure 23
Species-level squamate phylogeny continued (V).
Figure 24
Species-level squamate phylogeny continued (W).
Figure 25
Species-level squamate phylogeny continued (X).
Figure 26
Species-level squamate phylogeny continued (Y).
Figure 27
Species-level squamate phylogeny continued (Z).
Figure 28
Species-level squamate phylogeny continued (AA).
Similar articles
- The phylogeny of squamate reptiles (lizards, snakes, and amphisbaenians) inferred from nine nuclear protein-coding genes.
Vidal N, Hedges SB. Vidal N, et al. C R Biol. 2005 Oct-Nov;328(10-11):1000-8. doi: 10.1016/j.crvi.2005.10.001. Epub 2005 Oct 27. C R Biol. 2005. PMID: 16286089 - Resolving the phylogeny of lizards and snakes (Squamata) with extensive sampling of genes and species.
Wiens JJ, Hutter CR, Mulcahy DG, Noonan BP, Townsend TM, Sites JW Jr, Reeder TW. Wiens JJ, et al. Biol Lett. 2012 Dec 23;8(6):1043-6. doi: 10.1098/rsbl.2012.0703. Epub 2012 Sep 19. Biol Lett. 2012. PMID: 22993238 Free PMC article. - Molecular phylogenetics of squamata: the position of snakes, amphisbaenians, and dibamids, and the root of the squamate tree.
Townsend T, Larson A, Louis E, Macey JR. Townsend T, et al. Syst Biol. 2004 Oct;53(5):735-57. doi: 10.1080/10635150490522340. Syst Biol. 2004. PMID: 15545252 - [Genomic structure and sex determination in squamate reptiles].
Kichigin IG, Trifonov VA. Kichigin IG, et al. Tsitologiia. 2013;55(4):253-8. Tsitologiia. 2013. PMID: 23875459 Review. Russian. - The State of Squamate Genomics: Past, Present, and Future of Genome Research in the Most Speciose Terrestrial Vertebrate Order.
Gable SM, Mendez JM, Bushroe NA, Wilson A, Byars MI, Tollis M. Gable SM, et al. Genes (Basel). 2023 Jul 1;14(7):1387. doi: 10.3390/genes14071387. Genes (Basel). 2023. PMID: 37510292 Free PMC article. Review.
Cited by
- Discovery of a new species of the subgenus Japonigekko (Squamata, Gekkonidae, Gekko) from the Hengduan Mountains, southwestern China: the best Japonigekko mountaineer.
Ma S, Shi SC, Shen C, Chang LM, Jiang JP. Ma S, et al. Zookeys. 2024 Oct 17;1215:289-309. doi: 10.3897/zookeys.1215.125043. eCollection 2024. Zookeys. 2024. PMID: 39464300 Free PMC article. - First record of Isospora amphiboluri in the thorny devil, Moloch horridus.
Adriaanse K, Morgan T, Gasser RB, Koehler AV. Adriaanse K, et al. Int J Parasitol Parasites Wildl. 2024 Sep 5;25:100983. doi: 10.1016/j.ijppaw.2024.100983. eCollection 2024 Dec. Int J Parasitol Parasites Wildl. 2024. PMID: 39310796 Free PMC article. - The Phylogenetic Relationships of Major Lizard Families Using Mitochondrial Genomes and Selection Pressure Analyses in Anguimorpha.
Zhan L, Chen Y, He J, Guo Z, Wu L, Storey KB, Zhang J, Yu D. Zhan L, et al. Int J Mol Sci. 2024 Aug 2;25(15):8464. doi: 10.3390/ijms25158464. Int J Mol Sci. 2024. PMID: 39126033 Free PMC article. - Chromosome-level genome assembly and annotation of the Rhabdophis nuchalis (Hubei keelback).
Duan M, Yang S, Li X, Tang X, Cheng Y, Luo J, Wang J, Song H, Wang Q, Zhu GX. Duan M, et al. Sci Data. 2024 Aug 8;11(1):850. doi: 10.1038/s41597-024-03708-z. Sci Data. 2024. PMID: 39117633 Free PMC article. - Novel phylogenomic inference and 'Out of Asia' biogeography of cobras, coral snakes and their allies.
Weinell JL, Burbrink FT, Das S, Brown RM. Weinell JL, et al. R Soc Open Sci. 2024 Aug 7;11(8):240064. doi: 10.1098/rsos.240064. eCollection 2024 Aug. R Soc Open Sci. 2024. PMID: 39113776 Free PMC article.
References
- Uetz P. The Reptile Database. http://www.reptile-database.org/ Accessed December, 2012.
- Greene HW. Snakes: the Evolution of Mystery in Nature. Berkeley: University of California Press; 1997.
- Vitt LJ, Caldwell JP. Herpetology. 4. Burlington: Elsevier; 2009.
- Pianka ER, Vitt LJ. Lizards: Windows to the Evolution of Diversity. Berkeley: University of California Press; 2003.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources