Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26 - PubMed (original) (raw)
. 2013 Aug 8;500(7461):227-31.
doi: 10.1038/nature12328. Epub 2013 Jul 7.
Yawei Hu, Qihui Wang, Jianxun Qi, Feng Gao, Yan Li, Yanfang Zhang, Wei Zhang, Yuan Yuan, Jinku Bao, Buchang Zhang, Yi Shi, Jinghua Yan, George F Gao
Affiliations
- PMID: 23831647
- PMCID: PMC7095341
- DOI: 10.1038/nature12328
Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26
Guangwen Lu et al. Nature. 2013.
Abstract
The newly emergent Middle East respiratory syndrome coronavirus (MERS-CoV) can cause severe pulmonary disease in humans, representing the second example of a highly pathogenic coronavirus, the first being SARS-CoV. CD26 (also known as dipeptidyl peptidase 4, DPP4) was recently identified as the cellular receptor for MERS-CoV. The engagement of the MERS-CoV spike protein with CD26 mediates viral attachment to host cells and virus-cell fusion, thereby initiating infection. Here we delineate the molecular basis of this specific interaction by presenting the first crystal structures of both the free receptor binding domain (RBD) of the MERS-CoV spike protein and its complex with CD26. Furthermore, binding between the RBD and CD26 is measured using real-time surface plasmon resonance with a dissociation constant of 16.7 nM. The viral RBD is composed of a core subdomain homologous to that of the SARS-CoV spike protein, and a unique strand-dominated external receptor binding motif that recognizes blades IV and V of the CD26 β-propeller. The atomic details at the interface between the two binding entities reveal a surprising protein-protein contact mediated mainly by hydrophilic residues. Sequence alignment indicates, among betacoronaviruses, a possible structural conservation for the region homologous to the MERS-CoV RBD core, but a high variation in the external receptor binding motif region for virus-specific pathogenesis such as receptor recognition.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
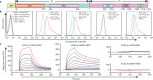
Figure 1. Identification of the MERS-CoV RBD.
a, A schematic representation of the MERS-CoV S protein. The N-terminal domain (NTD) and RBD are defined on the basis of a pairwise sequence alignment with the N-terminal galectin-like domain of murine hepatitis virus S and the RBD of SARS-CoV S, respectively. The remaining domain elements are bioinformatically defined on the basis of the web-server predictions (signal peptide (SP), SignalP 4.0 server; transmembrane domain (TM), TMHMM server; heptad repeats 1 and 2 (HR1 and HR2), Learncoil-VMF program). ? denotes the presumed/estimated S1/S2 cleavage site. A previous prediction indicates cleavage between R751 and S752 with a 602-residue S2. However, a recent study revealed a spike C-terminal domain (possibly S2) of ∼100 kDa, indicating a cleavage site upstream of R751/S752. b, A flow cytometric assay of the Fc-fused S protein or its subdomain proteins involved in CD26 binding. Mock-transfected baby hamster kidney (BHK) cells or BHK cells transfected with CD26-expressing plasmid (BHK-CD26) were tested with the individual Fc-fusion proteins or an anti-CD26 antibody (anti-CD26 IgG). For each test, the secondary antibodies (anti-goat IgG or anti-mouse IgG) were used as the negative control. The profiles are shown. From left to right: BHK cells with the indicated Fc-fusion proteins or antibodies, BHK-CD26 with anti-CD26 antibody, BHK-CD26 with Fc-fused S1, BHK-CD26 with Fc-fused NTD, BHK-CD26 with Fc-fused RBD. c, A surface plasmon resonance assay characterizing the specific binding between CD26 and MERS-CoV RBD. The profiles are shown. Left, human ACE2 to SARS-CoV RBD; middle, CD26 to MERS-CoV RBD; top right, CD26 to SARS-CoV RBD; bottom right, human ACE2 to MERS-CoV RBD. PowerPoint slide
Figure 2. The overall structure of MERS-CoV RBD.
a, A cartoon representation of the RBD structure. The secondary structural elements are labelled according to their occurrence in sequence. The disulphide bonds (marked with Arabic numbers 1–4) and N-glycan linked to N410 are shown as orange and green sticks, respectively. Core subdomain, magenta; external subdomain, cyan. The N and C termini are labelled. b, An amino acid sequence alignment between MERS-CoV and SARS-CoV RBDs. The hollow boxes and arrows indicate α/310 helices and β-strands, respectively, and are coloured as in a. To facilitate comparison, the secondary-structure elements of SARS-CoV RBD (PDB code, 2DD8) are marked with spiral (helices) and arrow (strands) lines below the sequence. The cysteine residues that form disulphide bonds are labelled as in a, and residue N410 with a star. c, A structural alignment between MERS-CoV (magenta for core and cyan for external subdomains) and SARS-CoV (green) RBDs. PowerPoint slide
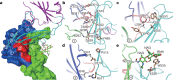
Figure 3. The complex structure of MERS-CoV RBD bound to CD26.
a, A cartoon representation of the complex structure. For clarity, only the β-propeller domain of CD26 (grey) is shown. Blades IV, V and the intervening IV/V linker that recognize RBD are highlighted in green, blue and red, respectively. The core subdomain and external RBM are coloured magenta and cyan, respectively. The right panel is yielded by clockwise rotation of the left panel along a longitudinal axis in the page-face. b, A symmetry-related CD26 dimer observed in the complex crystal. The two-fold axis is shown as an upright arrow. The transmembrane topology of CD26 is indicated with a modelled lipid-bilayer membrane. In CD26, the propeller and side openings indicated as the substrate entrance/exit tunnels are marked with arrows, and the catalytic triad residues are highlighted as spheres. Colour selections are the same as in a, and the CD26 α/β hydrolase domain is shown in orange. The N and C termini are labelled. PowerPoint slide
Figure 4. The atomic interaction details at the binding interface.
a, An overview of the binding interface. CD26 and RBD are shown in surface and cartoon representations, respectively, and are coloured as in Fig. 3. The carbohydrate moiety linked to CD26 N229 is shown as green sticks. The contacting sites (each allocated with an Arabic number 1–4) are further delineated in b–e for the amino acid interaction details. b, A strong polar-contact (H-bond and salt-bridge) network. c, d, The small patches of hydrophobic interactions. e, Contribution of the carbohydrate moiety. The residues involved are shown and labelled. NAG, _N_-acetyl-
D
-glucosamine; BMA, beta-
D
-mannose. PowerPoint slide
Comment in
- Spiking the MERS-coronavirus receptor.
Bosch BJ, Raj VS, Haagmans BL. Bosch BJ, et al. Cell Res. 2013 Sep;23(9):1069-70. doi: 10.1038/cr.2013.108. Epub 2013 Aug 13. Cell Res. 2013. PMID: 23938293 Free PMC article.
Similar articles
- Spread of Mutant Middle East Respiratory Syndrome Coronavirus with Reduced Affinity to Human CD26 during the South Korean Outbreak.
Kim Y, Cheon S, Min CK, Sohn KM, Kang YJ, Cha YJ, Kang JI, Han SK, Ha NY, Kim G, Aigerim A, Shin HM, Choi MS, Kim S, Cho HS, Kim YS, Cho NH. Kim Y, et al. mBio. 2016 Mar 1;7(2):e00019. doi: 10.1128/mBio.00019-16. mBio. 2016. PMID: 26933050 Free PMC article. - Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4.
Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Guo D, Fu L, Cui Y, Liu X, Arledge KC, Chen YH, Zhang L, Wang X. Wang N, et al. Cell Res. 2013 Aug;23(8):986-93. doi: 10.1038/cr.2013.92. Epub 2013 Jul 9. Cell Res. 2013. PMID: 23835475 Free PMC article. - Spiking the MERS-coronavirus receptor.
Bosch BJ, Raj VS, Haagmans BL. Bosch BJ, et al. Cell Res. 2013 Sep;23(9):1069-70. doi: 10.1038/cr.2013.108. Epub 2013 Aug 13. Cell Res. 2013. PMID: 23938293 Free PMC article. - The receptor binding domain of MERS-CoV: the dawn of vaccine and treatment development.
Zhou N, Zhang Y, Zhang JC, Feng L, Bao JK. Zhou N, et al. J Formos Med Assoc. 2014 Mar;113(3):143-7. doi: 10.1016/j.jfma.2013.11.006. Epub 2013 Dec 15. J Formos Med Assoc. 2014. PMID: 24342026 Free PMC article. Review. - A structural view of coronavirus-receptor interactions.
Reguera J, Mudgal G, Santiago C, Casasnovas JM. Reguera J, et al. Virus Res. 2014 Dec 19;194:3-15. doi: 10.1016/j.virusres.2014.10.005. Epub 2014 Oct 14. Virus Res. 2014. PMID: 25451063 Free PMC article. Review.
Cited by
- Emerging and reemerging infectious diseases: global trends and new strategies for their prevention and control.
Wang S, Li W, Wang Z, Yang W, Li E, Xia X, Yan F, Chiu S. Wang S, et al. Signal Transduct Target Ther. 2024 Sep 11;9(1):223. doi: 10.1038/s41392-024-01917-x. Signal Transduct Target Ther. 2024. PMID: 39256346 Free PMC article. Review. - Molecular basis of convergent evolution of ACE2 receptor utilization among HKU5 coronaviruses.
Park YJ, Liu C, Lee J, Brown JT, Ma CB, Liu P, Xiong Q, Stewart C, Addetia A, Craig CJ, Tortorici MA, Alshukari A, Starr T, Yan H, Veesler D. Park YJ, et al. bioRxiv [Preprint]. 2024 Aug 28:2024.08.28.608351. doi: 10.1101/2024.08.28.608351. bioRxiv. 2024. PMID: 39253417 Free PMC article. Preprint. - Potential Effects of Hyperglycemia on SARS-CoV-2 Entry Mechanisms in Pancreatic Beta Cells.
Michaels TM, Essop MF, Joseph DE. Michaels TM, et al. Viruses. 2024 Aug 2;16(8):1243. doi: 10.3390/v16081243. Viruses. 2024. PMID: 39205219 Free PMC article. Review. - Dissecting humoral immune responses to an MVA-vectored MERS-CoV vaccine in humans using a systems serology approach.
Weskamm LM, Tarnow P, Harms C, Huchon M, Raadsen MP, Friedrich M, Rübenacker L, Grüttner C, Garcia MG; MVA-MERS-S-CEF study group; Koch T, Becker S, Sutter G, Lhomme E, Haagmans BL, Fathi A, Blois SM, Dahlke C, Richert L, Addo MM. Weskamm LM, et al. iScience. 2024 Jul 8;27(8):110470. doi: 10.1016/j.isci.2024.110470. eCollection 2024 Aug 16. iScience. 2024. PMID: 39148710 Free PMC article. - The Chameleon Strategy-A Recipe for Effective Ligand Screening for Viral Targets Based on Four Novel Structure-Binding Strength Indices.
Latosińska M, Latosińska JN. Latosińska M, et al. Viruses. 2024 Jul 3;16(7):1073. doi: 10.3390/v16071073. Viruses. 2024. PMID: 39066235 Free PMC article.
References
- Bermingham A, et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17:20290. - PubMed
- World Health Organization. Cumulative Number of Reported Probable Cases of Severe Acute Respiratory Syndrome (SARS). http://www.who.int/csr/sars/country/en/
- World Health Organization. Novel coronavirus infection - update. http://www.who.int/csr/don/2013_05_15_ncov/en/ (2013)
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous