Molecular mechanisms underlying genotype-dependent responses to dietary restriction - PubMed (original) (raw)
. 2013 Dec;12(6):1050-61.
doi: 10.1111/acel.12130. Epub 2013 Aug 11.
Simon C Johnson, Christopher F Bennett, Marissa Simko, Natalie Trongtham, Anthony Castanza, Edward J Hsieh, Richard M Moller, Brian M Wasko, Joe R Delaney, George L Sutphin, Daniel Carr, Christopher J Murakami, Autumn Tocchi, Bo Xian, Weiyang Chen, Tao Yu, Sarani Goswami, Sean Higgins, Mollie Holmberg, Ki-Soo Jeong, Jin R Kim, Shannon Klum, Eric Liao, Michael S Lin, Winston Lo, Hillary Miller, Brady Olsen, Zhao J Peng, Tom Pollard, Prarthana Pradeep, Dillon Pruett, Dilreet Rai, Vanessa Ros, Minnie Singh, Benjamin L Spector, Helen Vander Wende, Elroy H An, Marissa Fletcher, Monika Jelic, Peter S Rabinovitch, Michael J MacCoss, Jing-Dong J Han, Brian K Kennedy, Matt Kaeberlein
Affiliations
- PMID: 23837470
- PMCID: PMC3838465
- DOI: 10.1111/acel.12130
Molecular mechanisms underlying genotype-dependent responses to dietary restriction
Jennifer Schleit et al. Aging Cell. 2013 Dec.
Abstract
Dietary restriction (DR) increases lifespan and attenuates age-related phenotypes in many organisms; however, the effect of DR on longevity of individuals in genetically heterogeneous populations is not well characterized. Here, we describe a large-scale effort to define molecular mechanisms that underlie genotype-specific responses to DR. The effect of DR on lifespan was determined for 166 single gene deletion strains in Saccharomyces cerevisiae. Resulting changes in mean lifespan ranged from a reduction of 79% to an increase of 103%. Vacuolar pH homeostasis, superoxide dismutase activity, and mitochondrial proteostasis were found to be strong determinants of the response to DR. Proteomic analysis of cells deficient in prohibitins revealed induction of a mitochondrial unfolded protein response (mtUPR), which has not previously been described in yeast. Mitochondrial proteotoxic stress in prohibitin mutants was suppressed by DR via reduced cytoplasmic mRNA translation. A similar relationship between prohibitins, the mtUPR, and longevity was also observed in Caenorhabditis elegans. These observations define conserved molecular processes that underlie genotype-dependent effects of DR that may be important modulators of DR in higher organisms.
Keywords: aging; dietary restriction; longevity; mitochondria; mitochondrial unfolded protein response; replicative lifespan; yeast.
© 2013 the Anatomical Society and John Wiley & Sons Ltd.
Figures
Figure 1. Genotype-dependent variation in yeast RLS response to dietary restriction
(A) RLS of 166 single deletion strains subjected to DR at 0.05% glucose. RLS varied from 80% reduction to 104% extension. Percent change in RLS on DR did not correlate to general fitness as measured by mean lifespan on 2% glucose (B) or division time (C). Gene ontology analysis of mutants with RLS significantly shortened (D) or extended (E) by DR revealed specific molecular processes involved in determining the direction of the response. Life span data and statistics provided in Table S1. GO analysis data provided in Tables S3-5.
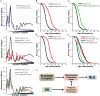
Figure 2. DR shortens the RLS of_sod2Δ_cells through the induction of respiratory metabolism
(A-B) Select mutants grown on DR demonstrate dose-dependency in both positive and negative effects of DR. (C) Growth of sod2Δ cells on respiratory media mimics the effect of DR on RLS. (D) Deletion of the transcription factor Hap4, which mediates the induction of respiration by DR, prevents the lifespan shortening effects of DR on sod2Δ cells. (E-F) RLS of sod2Δ and sod1Δ cells is increased on 2% and 0.05% glucose by addition of 20mM ascorbic acid. (G) sod2Δ cells are sensitive to the mitochondrial oxidative stress inducer paraquat and this sensitivity is increased under respiratory growth conditions. (H) This data supports a model by which respiratory growth induced by DR results in increased oxidative stress in sod2Δ cells, resulting in decreased RLS. Summary life span data are presented in Tables S7-8.
Figure 3. The DR mediated increase in RLS in_phbΔ_cells is not a result of increased respiration or autophagy and_phbΔ_cells show markers of mitochondrial proteotoxic stress which is attenuated by DR
(A) Inhibition of respiration, by deletion of HAP4 or RIP1 does not prevent the increased RLS resulting from DR in phbΔ cells. (B) Deletion of genes associated with autophagy does not prevent the RLS increase resulting from DR. (C) Growth on the non-fermentable carbon source, 3% glycerol, extends the RLS of phbΔ cell RLS to the same extent as 0.05% glucose. (D) Crude-mitochondrial isolation method schematic. (E) phbΔ cells show a significantly altered mitochondrial proteome when grown on 2% glucose but appear very similar to wild-type cells when grown in 3% glycerol. (F) Peptides annotated as mitochondrial were upregulated in phbΔ cells in 2% glucose. (G-H) Western blotting of whole cell protein isolates verifies induction of the mitochondrial chaperone Hsp-60 in phbΔ cells in 2% glucose and an attenuation of this in 3% glycerol. Summary life span data and proteomics are presented in Tables S1, S7-9.
Figure 4. Increased RLS of cells deficient in prohibitin by DR is mediated by decreased translation
(A) Polysome profiles of BY4742 cells on 2% glucose and 3% glycerol (top graph), phbΔ, rpl20bΔ, and phbΔ rpl20bΔ cells (middle graph), and BY4742, phbΔ, and sch9Δ phbΔ (bottom graph). (B) Deletion of the large ribosome subunit component Rpl20b increases RLS of phbΔ cells. (C) Deletion of GCN4 does not prevent the extensions of RLS in rpl20bΔ phbΔ cells. (D) Loss of Sch9 extends RLS of phbΔ cells to a an extent similar to DR. (E) Inhibition of translation by cycloheximide slightly, but significantly extends RLS of phbΔ cells. (F) Proposed model by which DR extends the RLS of phbΔ cells by reducing mitochondrial proteotoxic stress via reduced cytoplasmic translation. Summary life span data are presented in Tables S1 and S8.
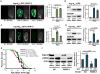
Figure 5. The effects of prohibitin deficiency on the mtUPR and response to DR are conserved in_C. elegans_
Fluorescent microscopy of _hsp-6pr∷_GFP (A) and _hsp-60pr∷_GFP (C) animals treated with empty vector (EV) (left panel), phb-2 (middle panel), or cco-1 (right panel) RNAi. Treatment with phb-2 and cco-1 RNAi induced expression of hsp-6 and hsp-60 (A). These findings were confirmed by Western blotting of whole protein extracts isolated hsp-6pr∷gfp and hsp-60pr∷gfp animals treated with EV, phb-2, or cco-1 RNAi (B, D). (E) phb-2 RNAi shortens the lifespan of wild-type animals, but extends the lifespan of animals lacking RSKS-1. Summary life span data and statistics are presented in Table S12. (F) Deletion of rsks-1 decreases basal levels and attenuates the induction of hsp-60 in animals fed phb-2 RNAi.
Similar articles
- Stress resistance and lifespan are increased in C. elegans but decreased in S. cerevisiae by mafr-1/maf1 deletion.
Cai Y, Wei YH. Cai Y, et al. Oncotarget. 2016 Mar 8;7(10):10812-26. doi: 10.18632/oncotarget.7769. Oncotarget. 2016. PMID: 26934328 Free PMC article. - d-Allulose, a stereoisomer of d-fructose, extends Caenorhabditis elegans lifespan through a dietary restriction mechanism: A new candidate dietary restriction mimetic.
Shintani T, Sakoguchi H, Yoshihara A, Izumori K, Sato M. Shintani T, et al. Biochem Biophys Res Commun. 2017 Dec 2;493(4):1528-1533. doi: 10.1016/j.bbrc.2017.09.147. Epub 2017 Sep 28. Biochem Biophys Res Commun. 2017. PMID: 28965946 - Dietary restriction induced longevity is mediated by nuclear receptor NHR-62 in Caenorhabditis elegans.
Heestand BN, Shen Y, Liu W, Magner DB, Storm N, Meharg C, Habermann B, Antebi A. Heestand BN, et al. PLoS Genet. 2013;9(7):e1003651. doi: 10.1371/journal.pgen.1003651. Epub 2013 Jul 25. PLoS Genet. 2013. PMID: 23935515 Free PMC article. - The mitochondrial unfolded protein response and increased longevity: cause, consequence, or correlation?
Bennett CF, Kaeberlein M. Bennett CF, et al. Exp Gerontol. 2014 Aug;56:142-6. doi: 10.1016/j.exger.2014.02.002. Epub 2014 Feb 8. Exp Gerontol. 2014. PMID: 24518875 Free PMC article. Review. - Dietary restriction and lifespan: Lessons from invertebrate models.
Kapahi P, Kaeberlein M, Hansen M. Kapahi P, et al. Ageing Res Rev. 2017 Oct;39:3-14. doi: 10.1016/j.arr.2016.12.005. Epub 2016 Dec 19. Ageing Res Rev. 2017. PMID: 28007498 Free PMC article. Review.
Cited by
- Promoting health and longevity through diet: from model organisms to humans.
Fontana L, Partridge L. Fontana L, et al. Cell. 2015 Mar 26;161(1):106-118. doi: 10.1016/j.cell.2015.02.020. Cell. 2015. PMID: 25815989 Free PMC article. Review. - Deficiency of the RNA-binding protein Cth2 extends yeast replicative lifespan by alleviating its repressive effects on mitochondrial function.
Patnaik PK, Beaupere C, Barlit H, Romero AM, Tsuchiya M, Muir M, Martínez-Pastor MT, Puig S, Kaeberlein M, Labunskyy VM. Patnaik PK, et al. Cell Rep. 2022 Jul 19;40(3):111113. doi: 10.1016/j.celrep.2022.111113. Cell Rep. 2022. PMID: 35858543 Free PMC article. - The aging lysosome: An essential catalyst for late-onset neurodegenerative diseases.
Nixon RA. Nixon RA. Biochim Biophys Acta Proteins Proteom. 2020 Sep;1868(9):140443. doi: 10.1016/j.bbapap.2020.140443. Epub 2020 May 13. Biochim Biophys Acta Proteins Proteom. 2020. PMID: 32416272 Free PMC article. Review. - Genomewide mechanisms of chronological longevity by dietary restriction in budding yeast.
Campos SE, Avelar-Rivas JA, Garay E, Juárez-Reyes A, DeLuna A. Campos SE, et al. Aging Cell. 2018 Jun;17(3):e12749. doi: 10.1111/acel.12749. Epub 2018 Mar 25. Aging Cell. 2018. PMID: 29575540 Free PMC article. - Age modifies respiratory complex I and protein homeostasis in a muscle type-specific manner.
Kruse SE, Karunadharma PP, Basisty N, Johnson R, Beyer RP, MacCoss MJ, Rabinovitch PS, Marcinek DJ. Kruse SE, et al. Aging Cell. 2016 Feb;15(1):89-99. doi: 10.1111/acel.12412. Epub 2015 Oct 25. Aging Cell. 2016. PMID: 26498839 Free PMC article.
References
- Artal-Sanz M, Tavernarakis N. Prohibitin couples diapause signalling to mitochondrial metabolism during ageing in C. elegans. Nature. 2009;461:793–797. - PubMed
- Artal-Sanz M, Tsang WY, Willems EM, Grivell LA, Lemire BD, van der Spek H, Nijtmans LG. The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans. The Journal of biological chemistry. 2003;278:32091–32099. - PubMed
- Coates PJ, Jamieson DJ, Smart K, Prescott AR, Hall PA. The prohibitin family of mitochondrial proteins regulate replicative lifespan. Curr Biol. 1997;7:607–610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 GM103533/GM/NIGMS NIH HHS/United States
- T32AG000057/AG/NIA NIH HHS/United States
- P01 AG001751/AG/NIA NIH HHS/United States
- P30AG013280/AG/NIA NIH HHS/United States
- P41GM103533/GM/NIGMS NIH HHS/United States
- T32ES007032/ES/NIEHS NIH HHS/United States
- T32 AG000057/AG/NIA NIH HHS/United States
- R01AG039390/AG/NIA NIH HHS/United States
- T32GM07270/GM/NIGMS NIH HHS/United States
- P30 AG013280/AG/NIA NIH HHS/United States
- R01 AG039390/AG/NIA NIH HHS/United States
- T32 ES007032/ES/NIEHS NIH HHS/United States
- T32 GM007270/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases