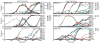Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations - PubMed (original) (raw)
. 2013 Aug 29;500(7464):571-4.
doi: 10.1038/nature12344. Epub 2013 Jul 21.
Affiliations
- PMID: 23873039
- PMCID: PMC3758440
- DOI: 10.1038/nature12344
Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations
Gregory I Lang et al. Nature. 2013.
Abstract
The dynamics of adaptation determine which mutations fix in a population, and hence how reproducible evolution will be. This is central to understanding the spectra of mutations recovered in the evolution of antibiotic resistance, the response of pathogens to immune selection, and the dynamics of cancer progression. In laboratory evolution experiments, demonstrably beneficial mutations are found repeatedly, but are often accompanied by other mutations with no obvious benefit. Here we use whole-genome whole-population sequencing to examine the dynamics of genome sequence evolution at high temporal resolution in 40 replicate Saccharomyces cerevisiae populations growing in rich medium for 1,000 generations. We find pervasive genetic hitchhiking: multiple mutations arise and move synchronously through the population as mutational 'cohorts'. Multiple clonal cohorts are often present simultaneously, competing with each other in the same population. Our results show that patterns of sequence evolution are driven by a balance between these chance effects of hitchhiking and interference, which increase stochastic variation in evolutionary outcomes, and the deterministic action of selection on individual mutations, which favours parallel evolutionary solutions in replicate populations.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Figure 1. The fates of individual spontaneously arising mutations
We show the frequency of all identified mutations through 1,000 generations in 6 of the 40 sequenced populations. Nonsynonymous mutations are solid lines with solid circles, while synonymous and intergenic mutations are dotted lines with open circles and squares respectively. Populations in the left and right columns were evolved at small (105) and large (106) population sizes, respectively. We observe qualitatively similar patterns in the other populations (Supplementary Fig. 1).
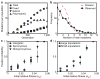
Figure 2. Statistical analysis across 40 replicate populations
a, The per-population number of total mutations, fixed mutations, extinct mutations, and mutations that are currently polymorphic over the course of the 1,000 generations. b, The distribution of the number of new mutations detected at each timepoint (solid blue line; see Methods for details) and a Poisson distribution with the same mean (dashed red line). c–d, Mutation fixation probability as a function of initial relative fitness. Data are mean±s.e.m.
Figure 3. The dynamics of sequence evolution in BYB1-G07
a, The trajectories of the 15 mutations that attain a frequency of at least 30%, hierarchically clustered into several distinct mutation “cohorts,” each of which is represented by a different color (Methods). b, Muller diagram showing the dynamics of the six main cohorts in the population. The number of times a mutation was observed in a given gene across all 40 populations is indicated in parentheses. Mutations in genes observed in more than three replicate populations (Table 1) are indicated in bold.
Figure 4. Genetic dissection of BYS1-A08
a, The trajectories of observed mutations. b, We crossed evolved clones from generation 545 to the ancestor; shown here are the fitnesses and genotypes of parental clones and 80 haploid progeny.
Similar articles
- mSphere of Influence: Deterministic and Stochastic Processes Drive Microbial Evolution.
Gerstein AC. Gerstein AC. mSphere. 2023 Apr 20;8(2):e0002223. doi: 10.1128/msphere.00022-23. Epub 2023 Feb 7. mSphere. 2023. PMID: 36749101 Free PMC article. - High-resolution lineage tracking reveals travelling wave of adaptation in laboratory yeast.
Nguyen Ba AN, Cvijović I, Rojas Echenique JI, Lawrence KR, Rego-Costa A, Liu X, Levy SF, Desai MM. Nguyen Ba AN, et al. Nature. 2019 Nov;575(7783):494-499. doi: 10.1038/s41586-019-1749-3. Epub 2019 Nov 13. Nature. 2019. PMID: 31723263 Free PMC article. - Identifying Targets of Selection in Laboratory Evolution Experiments.
Martínez AA, Lang GI. Martínez AA, et al. J Mol Evol. 2023 Jun;91(3):345-355. doi: 10.1007/s00239-023-10096-2. Epub 2023 Feb 21. J Mol Evol. 2023. PMID: 36810618 Free PMC article. Review. - Sex speeds adaptation by altering the dynamics of molecular evolution.
McDonald MJ, Rice DP, Desai MM. McDonald MJ, et al. Nature. 2016 Mar 10;531(7593):233-6. doi: 10.1038/nature17143. Epub 2016 Feb 24. Nature. 2016. PMID: 26909573 Free PMC article. - Genomic investigations of evolutionary dynamics and epistasis in microbial evolution experiments.
Jerison ER, Desai MM. Jerison ER, et al. Curr Opin Genet Dev. 2015 Dec;35:33-9. doi: 10.1016/j.gde.2015.08.008. Epub 2015 Sep 14. Curr Opin Genet Dev. 2015. PMID: 26370471 Free PMC article. Review.
Cited by
- The fitness consequences of aneuploidy are driven by condition-dependent gene effects.
Sunshine AB, Payen C, Ong GT, Liachko I, Tan KM, Dunham MJ. Sunshine AB, et al. PLoS Biol. 2015 May 26;13(5):e1002155. doi: 10.1371/journal.pbio.1002155. eCollection 2015 May. PLoS Biol. 2015. PMID: 26011532 Free PMC article. - Quantitative evolutionary dynamics using high-resolution lineage tracking.
Levy SF, Blundell JR, Venkataram S, Petrov DA, Fisher DS, Sherlock G. Levy SF, et al. Nature. 2015 Mar 12;519(7542):181-6. doi: 10.1038/nature14279. Epub 2015 Feb 25. Nature. 2015. PMID: 25731169 Free PMC article. - Synonymous Genetic Variation in Natural Isolates of Escherichia coli Does Not Predict Where Synonymous Substitutions Occur in a Long-Term Experiment.
Maddamsetti R, Hatcher PJ, Cruveiller S, Médigue C, Barrick JE, Lenski RE. Maddamsetti R, et al. Mol Biol Evol. 2015 Nov;32(11):2897-904. doi: 10.1093/molbev/msv161. Epub 2015 Jul 20. Mol Biol Evol. 2015. PMID: 26199375 Free PMC article. - A simple stochastic model describing the evolution of genomic GC content in asexually reproducing organisms.
Bohlin J. Bohlin J. Sci Rep. 2022 Nov 3;12(1):18569. doi: 10.1038/s41598-022-21709-z. Sci Rep. 2022. PMID: 36329129 Free PMC article. - Experimental Evolution of Metabolic Dependency in Bacteria.
D'Souza G, Kost C. D'Souza G, et al. PLoS Genet. 2016 Nov 4;12(11):e1006364. doi: 10.1371/journal.pgen.1006364. eCollection 2016 Nov. PLoS Genet. 2016. PMID: 27814362 Free PMC article.
References
- Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian Evolution Can Follow Only Very Few Mutational Paths to Fitter Proteins. Science. 2006;312:111–114. - PubMed
- Levin BR, Bull JJ. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends in Microbiology. 1994;2:76–81. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 GM046406/GM/NIGMS NIH HHS/United States
- R37 GM046406/GM/NIGMS NIH HHS/United States
- P50 GM071508/GM/NIGMS NIH HHS/United States
- GM046406/GM/NIGMS NIH HHS/United States
- GM071508/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases