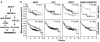RKIP and HMGA2 regulate breast tumor survival and metastasis through lysyl oxidase and syndecan-2 - PubMed (original) (raw)
RKIP and HMGA2 regulate breast tumor survival and metastasis through lysyl oxidase and syndecan-2
M Sun et al. Oncogene. 2014.
Abstract
Elucidating targets of physiological tumor metastasis suppressors can highlight key signaling pathways leading to invasion and metastasis. To identify downstream targets of the metastasis suppressor Raf-1 kinase inhibitory protein (RKIP/PEBP1), we utilized an integrated approach based upon statistical analysis of tumor gene expression data combined with experimental validation. Previous studies from our laboratory identified the architectural transcription factor and oncogene, high mobility group AT-hook 2 (HMGA2), as a target of inhibition by RKIP. Here we identify two signaling pathways that promote HMGA2-driven metastasis. Using both human breast tumor cells and an MMTV-Wnt mouse breast tumor model, we show that RKIP induces and HMGA2 inhibits expression of miR-200b; miR-200b directly inhibits expression of lysyl oxidase (LOX), leading to decreased invasion. RKIP also inhibits syndecan-2 (SDC2), which is aberrantly expressed in breast cancer, via downregulation of HMGA2; but this mechanism is independent of miR-200. Depletion of SDC2 induces apoptosis and suppresses breast tumor growth and metastasis in mouse xenografts. RKIP, LOX and SDC2 are coordinately regulated and collectively encompass a prognostic signature for metastasis-free survival in ER-negative breast cancer patients. Taken together, our findings reveal two novel signaling pathways targeted by the metastasis suppressor RKIP that regulate remodeling of the extracellular matrix and tumor survival.
Conflict of interest statement
Conflict Of Interest: The authors declare no conflict of interest.
Figures
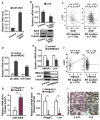
Figure 1
Bioinformatics scheme for selection of RKIP-mediated microRNA(s) and/or target gene(s) that may regulate breast tumor metastasis.
Figure 2
Expression of RKIP or depletion of HMGA2 induces miR-200b and inhibits LOX expression in 1833 cells, a bone-tropic derivative of the ER-negative human breast cancer cell line MDA-MB-231. (a,b) Expression of RKIP induced miR-200b and inhibited LOX expression. 1833 cells were stably transduced with S153E-RKIP or vector control. Samples were analyzed by qRT-PCR and immunoblotting for: (a) miR-200b RNA; (b) LOX mRNA (upper panel), and RKIP and LOX protein (lower panel). (c) Significant negative correlation between RKIP and LOX expression in both ER-negative (n = 175) and ER-positive (n = 556) breast cancer patient data (Br731). (d,e) Depletion of HMGA2 induced miR-200b and inhibited LOX expression. 1833 cells were stably transduced with HMGA2 shRNA (shHMGA2) or scrambled shRNA (control). Samples were analyzed by qRT-PCR and immunoblotting for: (d) primary miR-200b transcript (pri-200b) and mature RNA (miR-200b); (e) HMGA2 and LOX mRNA (upper panel) and protein (lower panel). (f) Significant positive correlation between HMGA2 and LOX expression observed in ER-negative (n = 175, left) but not in ER-positive (n = 556, right) breast cancer patient data (Br731). (g,h,i) Loss of Hmga2 in MMTV-Wnt1 transgenic mouse breast tumors induced miR-200b and inhibited LOX expression. Wnt1 transgenic mice were crossed with Hmga2 specific knockout mice. Mouse primary breast tumors were obtained from Hmga2 wildtype (Hmga2+/+) or null (Hmga2-/-) mice: (g) Murine miR-200b RNA and (h) Murine Hmga2 and Lox mRNA analyzed by qRT-PCR; (i) Hematoxylin and eosin (H&E) (Left) and murine Lox protein (Right) analyzed by immunostaining. (a,b,d,e,g,h) human or murine GAPDH was used as the normalization control for mRNA and U6 snRNA was used as the normalization control for microRNA; Tubulin was a loading control for protein; Data are mean ± s.e., n = 3. *, P < 0.05; **, P < 0.01. (c,f) Correlations were determined by Pearson's correlation coefficient. P value was determined by Student t test.
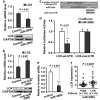
Figure 3
LOX is a direct target of miR-200b. (a) Transfection of 1833 cells with miR-200b precursor inhibited LOX expression: LOX mRNA was analyzed by qRT-PCR (upper panel) and protein by immunoblotting (lower panel). (b) Transfection of MCF-7 cells with anti-miR-200b induced LOX expression: LOX mRNA was analyzed by qRT-PCR (upper panel) and protein by immunoblotting (lower panel). (a,b) GAPDH as control. (c) Wildtype 3′UTR fragment in the LOX mRNA (LOX-wt-UTR) with the predicted binding site of miR-200b, and the mutant fragment (LOX-mut-UTR) showing the mutated sequence. (d) Luciferase and mutagenesis assay. LOX-wt-UTR and LOX-mut-UTR fragments were fused to a luciferase reporter (pMIR-REPORTER). Following co-transfection with miR-200b into cells, cell extracts were analyzed for luciferase activity as described in Methods. (e) Transfection of 1833 cells with miR-200b precursor inhibited cell invasion. Invasion assays were conducted as described in Methods. (a,b,d,e) Data are mean ± s.e., n = 3. (f) Significant negative correlation between miR-200b and LOX expression in breast cancer patients (Br86). P value is determined by Wilcoxon rank sum test.
Figure 4
Expression of RKIP or depletion of HMGA2 inhibits SDC2 expression. (a) RKIP inhibited SDC2 expression. 1833 cells were stably transduced with S153E-RKIP or vector control. Samples were analyzed by qRT-PCR for SDC2 mRNA (upper panel) or by immunoblotting for RKIP and SDC2 protein (lower panel). (b) Depletion of RKIP by RKIP shRNA induced HMGA2 and SDC2 expression. MDA-MB-435 cells were stably transduced with RKIP shRNA (shRKIP) or scrambled shRNA (Control). Samples were analyzed by qRT-PCR (upper panel) and immunoblotting (lower panel) for RKIP, HMGA2 and SDC2 expression. (c) Depletion of HMGA2 by HMGA2 shRNA inhibited SDC2 expression. 1833 cells were stably transduced with HMGA2 shRNA (shHMGA2) or scrambled shRNA (Control). Samples were analyzed by qRT-PCR (upper panel) and immunoblotting (lower panel) for HMGA2 and SDC2 expression. (d) Loss of Hmga2 in MMTV-Wnt1 transgenic mouse breast tumors inhibited SDC2 expression. Mouse primary breast tumors were obtained from MMTV-Wnt1/Hmga2+/+ or MMTV-Wnt1/Hmga2-/- mice. Murine Sdc2 protein was analyzed by immunostaining. (e) Significant positive correlation between HMGA2 and SDC2 expression observed in ER-negative (n = 175) (Left) but not in ER-positive (n = 556) (Right) breast cancer patient data (Br731). Correlations were determined by Pearson's correlation coefficient. P, Student t test. (a-c) GAPDH was used as the normalization control for mRNA; Tubulin was used as a loading control for protein. Data are mean ± s.e., n = 3. *, P < 0.05; **, P < 0.01.
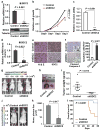
Figure 5
Depletion of SDC2 suppresses breast cancer cell growth, invasion, and metastasis. (a-c) Depletion of SDC2 inhibited cell growth and invasion in vitro. 1833 cells stably transduced with SDC2 shRNA (shSDC2) or scrambled shRNA (Control) were analyzed for: (a) SDC2 mRNA by qRT-PCR (upper panel) and protein by immunoblotting (lower panel); (b) Relative cell growth; or (c) Relative cell invasion. (d-i) Depletion of SDC2 suppressed breast tumor growth in vivo. 1833 cells stably transduced with shSDC2 or Control were orthotopically injected into the mammary fat pad of nude mice. Tumors were dissected at 6 weeks after implantation and analyzed: (d) by qRT-PCR for SDC2 mRNA; (e) by immunostaining for Hematoxylin and eosin (H&E) (Left) and SDC2 protein (Right); (f) by immunostaining for cleaved caspase-3. Representative images (Left) and quantification (Right) of apoptotic cells are shown; (g) Representative bioluminescence images of mice bearing 1833 cells treated as indicated; (h) Photograph of representative xenograft breast tumors of 1833 cells treated as indicated; (i) Xenograft breast tumors of 1833 cells treated as indicated and analyzed for tumor weight. Data are mean ± s.e., n = 7-10 per group. (j-l) Depletion of SDC2 suppressed breast tumor metastasis and enhanced overall survival. 1833 cells stably transduced with shSDC2 or Control were injected into the left ventricle of mice. Mice were imaged for luciferase activity after 3 weeks. (j) Representative bioluminescence images of mice with bone metastasis; (k) Quantification of bone colonization by 1833 cells treated as indicated. Data are mean ± s.e., n = 7-10 per group; (l) Kaplan-Meier survival analysis of mice over 8 weeks after injection of the tumor cells. (a-d) Data are mean ± s.e., n = 3.
Figure 6
The RKIP/LOX/SDC2 signaling pathways regulate breast tumorigenesis and stratify ER-negative subtype breast cancer patients for metastasis-free survival. (a) Scheme illustrating RKIP/HMGA2/miR-200b/LOX and RKIP/HMGA2/SDC2 signaling pathways in breast tumorigenesis. (b) Kaplan-Meier analysis of gene expression data from Br731 including 175 ER-negative and 556 ER-positive samples. Patients were stratified for metastasis-free survival in ER-negative or ER-positive patients using gene expression for RKIP, LOX, or SDC2 individually or the combined pathways as indicated. Right panel (RKIP/LOX/SDC2): Light gray line, high RKIP and low LOX/SDC2; Dark gray line, low RKIP and high LOX/SDC2. Top right panel: light gray line, n=22; dark gray line, n=46. P, chi-square p value.
Similar articles
- RKIP suppresses the proliferation and metastasis of breast cancer cell lines through up-regulation of miR-185 targeting HMGA2.
Zou Q, Wu H, Fu F, Yi W, Pei L, Zhou M. Zou Q, et al. Arch Biochem Biophys. 2016 Nov 15;610:25-32. doi: 10.1016/j.abb.2016.09.007. Epub 2016 Sep 17. Arch Biochem Biophys. 2016. PMID: 27651238 - Overexpression of RKIP inhibits cell invasion in glioma cell lines through upregulation of miR-98.
Chen Z, Cheng Q, Ma Z, Xi H, Peng R, Jiang B. Chen Z, et al. Biomed Res Int. 2013;2013:695179. doi: 10.1155/2013/695179. Epub 2013 Dec 12. Biomed Res Int. 2013. PMID: 24392454 Free PMC article. - Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer.
Yun J, Frankenberger CA, Kuo WL, Boelens MC, Eves EM, Cheng N, Liang H, Li WH, Ishwaran H, Minn AJ, Rosner MR. Yun J, et al. EMBO J. 2011 Aug 26;30(21):4500-14. doi: 10.1038/emboj.2011.312. EMBO J. 2011. PMID: 21873975 Free PMC article. - Inverse correlation between the metastasis suppressor RKIP and the metastasis inducer YY1: Contrasting roles in the regulation of chemo/immuno-resistance in cancer.
Wottrich S, Kaufhold S, Chrysos E, Zoras O, Baritaki S, Bonavida B. Wottrich S, et al. Drug Resist Updat. 2017 Jan;30:28-38. doi: 10.1016/j.drup.2017.01.001. Epub 2017 Jan 9. Drug Resist Updat. 2017. PMID: 28363333 Review. - Identification of novel metastasis suppressor signaling pathways for breast cancer.
Minn AJ, Bevilacqua E, Yun J, Rosner MR. Minn AJ, et al. Cell Cycle. 2012 Jul 1;11(13):2452-7. doi: 10.4161/cc.20624. Epub 2012 Jul 1. Cell Cycle. 2012. PMID: 22659842 Free PMC article. Review.
Cited by
- Cell surface heparan sulfate proteoglycans control adhesion and invasion of breast carcinoma cells.
Lim HC, Multhaupt HA, Couchman JR. Lim HC, et al. Mol Cancer. 2015 Jan 27;14(1):15. doi: 10.1186/s12943-014-0279-8. Mol Cancer. 2015. PMID: 25623282 Free PMC article. - The Significance of Human Papillomavirus Receptors Related Genetic Variants in Cervical Cancer Screening.
Xie H, Wei M, Yao L, Liu Y, Xie X, Li X. Xie H, et al. Microbiol Spectr. 2023 Aug 17;11(4):e0511722. doi: 10.1128/spectrum.05117-22. Epub 2023 Jun 26. Microbiol Spectr. 2023. PMID: 37358427 Free PMC article. - Robust performance of a novel stool DNA test of methylated SDC2 for colorectal cancer detection: a multicenter clinical study.
Wang J, Liu S, Wang H, Zheng L, Zhou C, Li G, Huang R, Wang H, Li C, Fan X, Fu X, Wang X, Guo H, Guan J, Sun Y, Song X, Li Z, Mu D, Sun J, Liu X, Qi Y, Niu F, Chen C, Wu X, Wang X, Song X, Zou H. Wang J, et al. Clin Epigenetics. 2020 Oct 30;12(1):162. doi: 10.1186/s13148-020-00954-x. Clin Epigenetics. 2020. PMID: 33126908 Free PMC article. Clinical Trial. - HMGA2 is associated with epithelial-mesenchymal transition and can predict poor prognosis in nasopharyngeal carcinoma.
Xia YY, Yin L, Tian H, Guo WJ, Jiang N, Jiang XS, Wu J, Chen M, Wu JZ, He X. Xia YY, et al. Onco Targets Ther. 2015 Jan 17;8:169-76. doi: 10.2147/OTT.S74397. eCollection 2015. Onco Targets Ther. 2015. PMID: 25653540 Free PMC article. - EDIL3 promotes epithelial-mesenchymal transition and paclitaxel resistance through its interaction with integrin αVβ3 in cancer cells.
Gasca J, Flores ML, Jiménez-Guerrero R, Sáez ME, Barragán I, Ruíz-Borrego M, Tortolero M, Romero F, Sáez C, Japón MA. Gasca J, et al. Cell Death Discov. 2020 Sep 16;6:86. doi: 10.1038/s41420-020-00322-x. eCollection 2020. Cell Death Discov. 2020. PMID: 33014430 Free PMC article.
References
- Massague J. Sorting out breast-cancer gene signatures. N Engl J Med. 2007;356:294–297. - PubMed
- Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. - PubMed
- Fu Z, Smith PC, Zhang L, Rubin MA, Dunn RL, Yao Z, et al. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J Natl Cancer Inst. 2003;95:878–889. - PubMed
- Fu Z, Kitagawa Y, Shen R, Shah R, Mehra R, Rhodes D, et al. Metastasis suppressor gene Raf kinase inhibitor protein (RKIP) is a novel prognostic marker in prostate cancer. Prostate. 2006;66:248–256. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous