First insights into the entry process of hyperthermophilic archaeal viruses - PubMed (original) (raw)
First insights into the entry process of hyperthermophilic archaeal viruses
Emmanuelle R J Quemin et al. J Virol. 2013 Dec.
Abstract
A decisive step in a virus infection cycle is the recognition of a specific receptor present on the host cell surface, subsequently leading to the delivery of the viral genome into the cell interior. Until now, the early stages of infection have not been thoroughly investigated for any virus infecting hyperthermophilic archaea. Here, we present the first study focusing on the primary interactions between the archaeal rod-shaped virus Sulfolobus islandicus rod-shaped virus 2 (SIRV2) (family Rudiviridae) and its hyperthermoacidophilic host, S. islandicus. We show that SIRV2 adsorption is very rapid, with the majority of virions being irreversibly bound to the host cell within 1 min. We utilized transmission electron microscopy and whole-cell electron cryotomography to demonstrate that SIRV2 virions specifically recognize the tips of pilus-like filaments, which are highly abundant on the host cell surface. Following the initial binding, the viral particles are found attached to the sides of the filaments, suggesting a movement along these appendages toward the cell surface. Finally, we also show that SIRV2 establishes superinfection exclusion, a phenomenon not previously described for archaeal viruses.
Figures
Fig 1
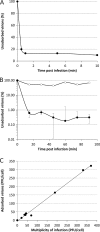
Adsorption of SIRV2 to cells of S. islandicus LAL14/1. (A) Kinetics of SIRV2 adsorption. Cells were infected with SIRV2 using an MOI of 0.1 at 75°C. The number of unbound virus particles was determined at different time points postinfection as described in Materials and Methods. (B) SIRV2-mediated superinfection exclusion. Cells were infected at an MOI of 10 for 1 h, washed twice with rich medium, and subjected to a second round of infection using an MOI of 0.1. The number of unadsorbed particles remaining in the supernatant was determined. The kinetics of adsorption to noninfected, control cells is shown by closed circles, while open circles represent adsorption to preinfected cells. All experiments were conducted in triplicate, and error bars represent standard deviations. When error bars are not visible, the deviation was below 5%. (C) Receptor saturation. Cells were infected with SIRV2 using MOIs ranging from 0.1 to 370. At 30 min postinfection, the number of unadsorbed viral particles present in the supernatant was determined by plaque assay and compared to that of the cell-free control.
Fig 2
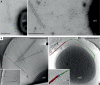
Electron micrographs of SIRV2 interaction with S. islandicus LAL14/1 cells. Samples were collected 1 min postinfection and negatively stained for TEM (A) or plunge-frozen for electron cryotomography (cryo-ET) (B). The virions interact both at the filament tips (right panels) and along the length of the filaments (left panels). The inset in the lower left panel depicts two virions bound to the sides of a single filament. The lower right panel shows a segmented tomographic volume of the SIRV2 virion (red) attached to the tip of an S. islandicus filament (green). The three terminal virion fibers that appear to mediate the interaction are shown in blue (the inset depicts a magnified view of the interaction between the virion fibers and the tip of the filament). A complete tomogram of the cell depicted in the lower right panel can be found in the supplemental material. Scale bars, 500 nm.
Fig 3
Transmission electron micrographs of SIRV2 interaction with purified cellular filaments. The filaments were removed from S. islandicus LAL14/1 cells as described in Materials and Methods.
Fig 4
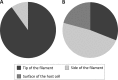
Interactions between SIRV2 and purified filaments (A) or S. islandicus LAL14/1 cells (B). SIRV2 virions were incubated with purified filaments for 5 to 10 min or LAL14/1 cells for 1 to 2 min at 75°C and prepared for TEM (see Materials and Methods). Binding of viral particles to the tips, the sides of LAL14/1 filaments, or the cell surface was counted in electron micrographs of negatively stained samples.
Fig 5
A tomographic slice through S. islandicus LAL14/1 cells 1 min after infection with SIRV2 reveals partially disassembled SIRV2 virions at the cell surface. Scale bar, 100 nm.
Similar articles
- DNA-Interacting Characteristics of the Archaeal Rudiviral Protein SIRV2_Gp1.
Peeters E, Boon M, Rollie C, Willaert RG, Voet M, White MF, Prangishvili D, Lavigne R, Quax TEF. Peeters E, et al. Viruses. 2017 Jul 18;9(7):190. doi: 10.3390/v9070190. Viruses. 2017. PMID: 28718834 Free PMC article. - Protein-protein interactions leading to recruitment of the host DNA sliding clamp by the hyperthermophilic Sulfolobus islandicus rod-shaped virus 2.
Gardner AF, Bell SD, White MF, Prangishvili D, Krupovic M. Gardner AF, et al. J Virol. 2014 Jun;88(12):7105-8. doi: 10.1128/JVI.00636-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696494 Free PMC article. - Virology. A virus that infects a hyperthermophile encapsidates A-form DNA.
DiMaio F, Yu X, Rensen E, Krupovic M, Prangishvili D, Egelman EH. DiMaio F, et al. Science. 2015 May 22;348(6237):914-7. doi: 10.1126/science.aaa4181. Science. 2015. PMID: 25999507 Free PMC article. - Genomics and biology of Rudiviruses, a model for the study of virus-host interactions in Archaea.
Prangishvili D, Koonin EV, Krupovic M. Prangishvili D, et al. Biochem Soc Trans. 2013 Feb 1;41(1):443-50. doi: 10.1042/BST20120313. Biochem Soc Trans. 2013. PMID: 23356326 Free PMC article. Review. - Exceptional virion release mechanism: one more surprise from archaeal viruses.
Prangishvili D, Quax TE. Prangishvili D, et al. Curr Opin Microbiol. 2011 Jun;14(3):315-20. doi: 10.1016/j.mib.2011.04.006. Epub 2011 Apr 30. Curr Opin Microbiol. 2011. PMID: 21531608 Review.
Cited by
- Surface resistance to SSVs and SIRVs in pilin deletions of Sulfolobus islandicus.
Rowland EF, Bautista MA, Zhang C, Whitaker RJ. Rowland EF, et al. Mol Microbiol. 2020 Apr;113(4):718-727. doi: 10.1111/mmi.14435. Epub 2019 Dec 19. Mol Microbiol. 2020. PMID: 31774609 Free PMC article. - Unique architecture of thermophilic archaeal virus APBV1 and its genome packaging.
Ptchelkine D, Gillum A, Mochizuki T, Lucas-Staat S, Liu Y, Krupovic M, Phillips SEV, Prangishvili D, Huiskonen JT. Ptchelkine D, et al. Nat Commun. 2017 Nov 10;8(1):1436. doi: 10.1038/s41467-017-01668-0. Nat Commun. 2017. PMID: 29127347 Free PMC article. - The enigmatic archaeal virosphere.
Prangishvili D, Bamford DH, Forterre P, Iranzo J, Koonin EV, Krupovic M. Prangishvili D, et al. Nat Rev Microbiol. 2017 Nov 10;15(12):724-739. doi: 10.1038/nrmicro.2017.125. Nat Rev Microbiol. 2017. PMID: 29123227 Review. - Diversity of Bathyarchaeia viruses in metagenomes and virus-encoded CRISPR system components.
Duan C, Liu Y, Liu Y, Liu L, Cai M, Zhang R, Zeng Q, Koonin EV, Krupovic M, Li M. Duan C, et al. ISME Commun. 2024 Jan 10;4(1):ycad011. doi: 10.1093/ismeco/ycad011. eCollection 2024 Jan. ISME Commun. 2024. PMID: 38328448 Free PMC article. - Archaeal extrachromosomal genetic elements.
Wang H, Peng N, Shah SA, Huang L, She Q. Wang H, et al. Microbiol Mol Biol Rev. 2015 Mar;79(1):117-52. doi: 10.1128/MMBR.00042-14. Microbiol Mol Biol Rev. 2015. PMID: 25694123 Free PMC article. Review.
References
- Krupovic M, White MF, Forterre P, Prangishvili D. 2012. Postcards from the edge: structural genomics of archaeal viruses. Adv. Virus Res. 82:33–62 - PubMed
- Pina M, Bize A, Forterre P, Prangishvili D. 2011. The archeoviruses. FEMS Microbiol. Rev. 35:1035–1054 - PubMed
- Prangishvili D. 2013. The wonderful world of archaeal viruses. Annu. Rev. Microbiol. 67:565–585 - PubMed
- Prangishvili D, Garrett RA, Koonin EV. 2006. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 117:52–67 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources