Salmonella modulation of host cell gene expression promotes its intracellular growth - PubMed (original) (raw)
Salmonella modulation of host cell gene expression promotes its intracellular growth
Sebastian Hannemann et al. PLoS Pathog. 2013.
Abstract
Salmonella Typhimurium has evolved a complex functional interface with its host cell largely determined by two type III secretion systems (T3SS), which through the delivery of bacterial effector proteins modulate a variety of cellular processes. We show here that Salmonella Typhimurium infection of epithelial cells results in a profound transcriptional reprogramming that changes over time. This response is triggered by Salmonella T3SS effector proteins, which stimulate unique signal transduction pathways leading to STAT3 activation. We found that the Salmonella-stimulated changes in host cell gene expression are required for the formation of its specialized vesicular compartment that is permissive for its intracellular replication. This study uncovers a cell-autonomous process required for Salmonella pathogenesis potentially opening up new avenues for the development of anti-infective strategies that target relevant host pathways.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
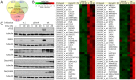
Figure 1. Salmonella Typhimurium triggers a complex gene expression program in cultured Henle-407 cells.
(A) Venn diagram depicting the number of unique and common genes whose expression changed at least 3 fold at the indicated times after S. Typhimurium infection. (B) Heat map of selected genes whose expression changed at least 12 fold at the indicated times after S. Typhimurium infection. (C) Western blot detection of selected proteins whose genes were significantly upregulated after S. Typhimurium infection. Henle-407 cells were infected (MOI = 10) with wild-type S. Typhimurium or the SPI-1 T3SS-defective ΔinvA mutant strain for 1 h. At the indicated times, cells were harvested, their lysates separated by SDS-PAGE and probed by immuno blotting with the specified antibodies to the indicated proteins of interest and to tubulin as a loading control. (n. i.: non infected).
Figure 2. Salmonella stimulation of transcriptional responses in infected cells requires STAT3.
(A) Interaction map of genes whose expression is stimulated by S. Typhimurium infection of Henle-407 cells. Shown is the interaction of genes whose expression increased at least 3 fold at 10 or 20 h after infection. The analysis was carried out with STRING 9.0 (
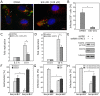
) using the highest confidence (0.9) parameters. (B–E) S. Typhimurium induces STAT3 activation. Henle-407 cells were infected (MOI = 10) with wild-type S. Typhimurium for 1 h. Following chase in gentamicin containing medium, cells were lysed at the indicated times, separated by SDS-PAGE and probed by immuno blotting with antibodies to the phosphorylated (activated) form of STAT3 (P-Y705), unphosphorylated STAT3 (to detect total amount), and actin (loading control) (B). Fold activation of STAT3 (relative to uninfected cells) was quantified by scanning the blots with a LI-COR Odyssey imaging system standardizing the signals with the actin loading control. Values are the means (± SD) of three independent experiments (C). Alternatively, Henle-407 cells were infected (MOI = 5) for 1 h with wild-type S. Typhimurium, chased for 8 h in gentamicin supplemented medium, fixed, immuno stained for LPS (red), DNA (blue) and phosphorylated STAT3 (green), as indicated. Samples were then analyzed by epifluorescence microscopy (bar represents 10 µm) (D). The number of infected cells showing staining of phosphorylated STAT3 was determined and the values represent the mean (± SD) of three independent experiments in which at least 100 cells were quantified. *: indicates values that are statistically significantly different from uninfected controls (_p_≤0.01) (E). (F) STAT3 is required for the transcriptional responses to S. Typhimurium infection. Henle-407 cells treated with the STAT3 inhibitor S31-201 were infected (MOI = 10) with S. Typhimurium for 1 h, and chased for additional 3 h in gentamicin containing medium in the presence of DMSO or 100 µM S3I-201. mRNA levels of selected genes whose expression is induced by S. Typhimurium infection was analyzed by qRT-PCR. Values represent the mean (± SEM) of GAPDH normalized transcript levels of the indicated genes, relative to DMSO treated uninfected cells. *: indicates statistically significant differences (_p_≤0.0004). (n. i.: non infected).
Figure 3. Salmonella stimulation of transcriptional responses in infected cells requires the SPI-1 T3SS effectors SopE, SopE2, and SopB.
(A) Salmonella induces STAT3 activation in a flagellin-independent manner. Cultured Henle-407 cells were infected with wild-type S. Typhimurium (MOI = 10) or a ΔflhD mutant strain (which does not express flagella) (MOI = 100) for 1 h, resulting in an equal number of infected cells (Note: higher MOI was used for the flagellar mutant to compensate for its reduced ability to infect cells). Infected cells were then chased in the presence of gentamicin, lysed at the indicated times, separated by SDS-PAGE and probed by immunoblotting with antibodies to phosphorylated (activated) form of STAT3 (P-Y705), and tubulin (as loading control). (B–D) Henle-407 cells were infected (MOI = 10) for 1 h with wild-type S. Typhimurium, the SPI-1 T3SS-deficient ΔinvA mutant (B), or mutants defective in the effectors sopE, sopE2, and/or sopB, (C) or the indicated mutants carrying a plasmid encoding the Yersinia pseudotuberculosis invasin protein (D). Following chase in gentamicin containing medium, cells were lysed at the indicated times, separated by SDS-PAGE and probed by immuno blotting with antibodies to the phosphorylated (activated) form of STAT3 (P-Y705), unphosphorylated STAT3 (to detect total amount), and tubulin (loading control). (n. i.: non infected).
Figure 4. Salmonella Typhimurium activates STAT3 by a non-canonical pathway that requires Abl and PAK.
(A) Culture supernatants from Henle-407 infected cells do not activate STAT3. Culture supernatants were obtained from Henle-407 cells 20 h after infection (MOI = 10) with either wild-type S. Typhimurium or the SPI-1 T3SS-defective ΔinvA mutant, filtered sterilized, and applied to uninfected Henle-407 cells. At different times after treatment cells were lysed, separated by SDS-PAGE and probed by immuno blotting with antibodies to the phosphorylated (activated) form of STAT3 (P-Y705), and tubulin (loading control). As a control, infected cells were analyzed for STAT3 activation in a similar fashion. (B) Activation of STAT3 by S. Typhimurium is JAK independent. Henle-407 cells were treated with the JAK inhibitor Tofacitinib at the indicated concentration and then infected (MOI = 10) with wild-type S. Typhimurium for 1 h, chased for additional 6 h in gentamicin containing medium in the presence of DMSO or Tofacitinib. Cells were then lysed and analyzed for STAT3 activation as indicated in (A). (C) Abl kinases are required for efficient S. Typhimurium-induced STAT3 activation. Cultured epithelial cells were pretreated with increasing concentrations of the STAT3 inhibitor Imatinib for 1 h, infected (MOI = 10) with wild-type S. Typhimurium, chased in the presence of the inhibitor, and at the indicated time cells were lysed and analyzed for STAT3 activation as described in (A). (D) S. Typhimurium-induced STAT3 activation requires PAK activity. Cultured epithelial cells were pretreated with increasing concentrations of the PAK inhibitor IPA-3 for 1 h, infected (MOI = 10) with wild-type S. Typhimurium, chased in the presence of the inhibitor, and at the indicated times cells were lysed and analyzed from STAT3 activation as indicated in (A). (E) Abl and PAK are required for the transcriptional responses to S. Typhimurium infection. Henle-407 cells were treated with inhibitors for Abl kinases (imatinib), PAK (IPA-3), JAK (Tofacitinib), and Src (PP1), infected (MOI = 10) with wild-type S. Typhimurium for 1 h, and chased for additional 3 h in gentamicin containing medium in the presence of DMSO or the inhibitors. mRNA levels of the selected indicated genes whose expression increase after S. Typhimurium infection were analyzed by qRT-PCR. Values represent the mean (± SEM) of GAPDH normalized transcript levels in infected cells relative to DMSO treated uninfected cells. *: indicates statistically significant differences (_p_≤0.02) of the indicated comparisons. (n. i.: non infected).
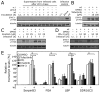
Figure 5. Host cell gene expression reprogramming is required for Salmonella intracellular fitness and replication.
(A) and (B) STAT3 activity is required for the formation of S. Typhimurium-induced filaments (SIFs). Henle-407 cells treated with the STAT3 inhibitor S31-201 (100 µM) or DMSO were infected (MOI = 5) with wild-type S. Typhimurium for 1 h, chased for additional 8 h in gentamicin-containing medium, in the presence of the inhibitor or DMSO. Cells were then fixed, immuno stained for LAMP1 (red) and LPS (green) and the number of infected cells showing the presence of SIFs enumerated by epifluorescence microscopy (arrows denote the presence of SIFs). Values are the mean percentages (± SD) of infected cells showing SIF formation and represent three independent experiments in which at least 100 cells per condition were examined. *: indicates statistically significant differences (_p_≤0.005). (C–E) STAT3 activity is required for intracellular bacterial growth. Henle-407 cells were treated with the STAT3 inhibitor S31-201 (100 µM) or DMSO and then infected (MOI = 5) with wild-type S. Typhimurium for 1 h, chased for the indicated times in the presence of gentamicin and the inhibitor, and intracellular c.f.u. was enumerated by plating dilutions in the appropriate media (c. f. u. at time 0.5 h were as follows: DMSO treated: 7.6×105±3.2×105; S31-201 treated: 9.6×105±4.9×105 (C). Alternatively, S. Typhimurium growth was examined in HeLa cells transfected with an shRNA construct targeting STAT3 following the same procedure (c. f. u. at time 0.5 h were as follows: vector treated: 6.6×105±2.4×105; STAT3 shRNA treated: 7.3×105±1.8×105 (D). Numbers represent fold replication and are the mean (± SD) of three independent determinations (C and D). *: indicates statistically significant differences (_p_≤0.007). Levels of STAT3 in shRNA-targeted cells were determined by immunoblotting with the indicated antibodies (E). (F) S. Typhimurium shows increased fitness and greater replication in SerpinB3-positive (i. e. transcriptionally reprogrammed) cells. Henle-407 cells were infected (MOI = 10) for 1 h with S. Typhimurium encoding dsRed under the control of an arabinose inducible promoter, chased for 17 h in gentamicin supplemented medium, and further incubated for 3 h in the presence of 0.1% arabinose. Cells were then fixed, immuno stained for LPS, endogenous SerpinB3, and DNA, and bacteria expressing dsRed in SerpinB3-positive or negative cells were enumerated by epifluorescence microscopy. Fitness was measured by evaluating the ability of intracellular bacteria to produce dsRed protein. Values are the means (± SD) of the percentages of the bacteria showing dsRed expression after addition of arabinose in SerpinB3-positive or negative cells, and represent data from three independent experiments in which at least 100 infected cells were examined (F) (*: _p_≤0.007). (G) Increased _Salmonella_-induced filament (SIFs) formation in SerpinB3-positive (i. e. transcriptionally reprogrammed) cells. Cultured epithelial cells were infected (MOI = 10) for 1 h with S. Typhimurium, chased for 20 h in gentamicin-supplemented medium, fixed, immuno stained for LAMP1 (to stain for SIFs), LPS and SerpinB3, and analyzed by epifluorescence microscopy. Depicted are the mean percentages (± SD) of infected, SerpinB3-positive cells that show the presence (SIF+) or absence (SIF−) of _Salmonella_-induced filaments (SIFs) from three independent experiments in which at least 100 cells per condition were analyzed (*: _p_≤0.001). (H) Increased bacterial replication in SerpinB3-positive cells. Cells were infected with wild type S. Typhimurium, (MOI = 10) chased for 20 h in gentamicin supplemented medium, fixed, immuno stained for LPS (to stain for Salmonella), endogenous SerpinB3 and DNA, and the total number of bacteria in SerpinB3-positive or negative cells were enumerated by epifluorescence microscopy. Values are the means (± SD) of the percentages of SerpinB3-positive or negative cells that had a bacterial load of up to 10 bacteria or more than 10 bacteria, and represent three independent experiments in which at least 100 cells per condition were examined (*: _p_≤0.00002).
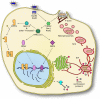
Figure 6. Bacterially-induced reprogramming of host cell gene expression is required for efficient Salmonella replication.
S. Typhimurium infects intestinal epithelial cells by delivering effector proteins into the host cell, using its SPI-1 T3SS. The effector proteins SopB, SopE and SopE2 activate the small GTPases Cdc42, Rac1 and RhoG in a redundant manner, thus inducing membrane ruffling and bacterial uptake. Internalized bacteria reside in macropinosomes, which then maturate into Salmonella containing vacuoles (SCVs). Bacterial effector-stimulated small GTPases also activate members of the p21 activated kinase family (PAK). These serine/threonine kinases trigger downstream signaling of mitogen-activated protein kinases (MAPKs) and nuclear factor kappa B (NFκB) , , , resulting in activation of additional transcription factors. In addition, activated PAK proteins also phosphorylate members of the Abl kinase family, thereby triggering auto-phosphorylation of Abl proteins for full activation, subsequently leading to the activation the cytoplasmic transcription factor STAT3. Products of STAT3-controlled genes ultimately influence vesicular trafficking between cellular compartments and the SCV, contributing to the formation of Salmonella induced filaments (SIFs), which characterize a fully mature, replication-competent _Salmonella_-containing vacuole.
Similar articles
- Tumor Necrosis Factor Receptor-Associated Factor 6 (TRAF6) Mediates Ubiquitination-Dependent STAT3 Activation upon Salmonella enterica Serovar Typhimurium Infection.
Ruan HH, Zhang Z, Wang SY, Nickels LM, Tian L, Qiao JJ, Zhu J. Ruan HH, et al. Infect Immun. 2017 Jul 19;85(8):e00081-17. doi: 10.1128/IAI.00081-17. Print 2017 Aug. Infect Immun. 2017. PMID: 28507064 Free PMC article. - Salmonella Activation of STAT3 Signaling by SarA Effector Promotes Intracellular Replication and Production of IL-10.
Jaslow SL, Gibbs KD, Fricke WF, Wang L, Pittman KJ, Mammel MK, Thaden JT, Fowler VG Jr, Hammer GE, Elfenbein JR, Ko DC. Jaslow SL, et al. Cell Rep. 2018 Jun 19;23(12):3525-3536. doi: 10.1016/j.celrep.2018.05.072. Cell Rep. 2018. PMID: 29924996 Free PMC article. - Deletion of invH gene in Salmonella enterica serovar Typhimurium limits the secretion of Sip effector proteins.
Pati NB, Vishwakarma V, Jaiswal S, Periaswamy B, Hardt WD, Suar M. Pati NB, et al. Microbes Infect. 2013 Jan;15(1):66-73. doi: 10.1016/j.micinf.2012.10.014. Epub 2012 Nov 13. Microbes Infect. 2013. PMID: 23159244 - Salmonellae interactions with host processes.
LaRock DL, Chaudhary A, Miller SI. LaRock DL, et al. Nat Rev Microbiol. 2015 Apr;13(4):191-205. doi: 10.1038/nrmicro3420. Epub 2015 Mar 9. Nat Rev Microbiol. 2015. PMID: 25749450 Free PMC article. Review. - Salmonella effectors: important players modulating host cell function during infection.
Agbor TA, McCormick BA. Agbor TA, et al. Cell Microbiol. 2011 Dec;13(12):1858-69. doi: 10.1111/j.1462-5822.2011.01701.x. Epub 2011 Oct 10. Cell Microbiol. 2011. PMID: 21902796 Free PMC article. Review.
Cited by
- The Salmonella Effector Protein SopA Modulates Innate Immune Responses by Targeting TRIM E3 Ligase Family Members.
Kamanova J, Sun H, Lara-Tejero M, Galán JE. Kamanova J, et al. PLoS Pathog. 2016 Apr 8;12(4):e1005552. doi: 10.1371/journal.ppat.1005552. eCollection 2016 Apr. PLoS Pathog. 2016. PMID: 27058235 Free PMC article. - Subverting Host Cell P21-Activated Kinase: A Case of Convergent Evolution across Pathogens.
John Von Freyend S, Kwok-Schuelein T, Netter HJ, Haqshenas G, Semblat JP, Doerig C. John Von Freyend S, et al. Pathogens. 2017 Apr 21;6(2):17. doi: 10.3390/pathogens6020017. Pathogens. 2017. PMID: 28430160 Free PMC article. Review. - Salmonella enterica serovar-specific transcriptional reprogramming of infected cells.
Hannemann S, Galán JE. Hannemann S, et al. PLoS Pathog. 2017 Jul 24;13(7):e1006532. doi: 10.1371/journal.ppat.1006532. eCollection 2017 Jul. PLoS Pathog. 2017. PMID: 28742135 Free PMC article. - Rapid pathogen-specific recruitment of immune effector cells in the skin by secreted toxins.
Nguyen TH, Cheung GYC, Rigby KM, Kamenyeva O, Kabat J, Sturdevant DE, Villaruz AE, Liu R, Piewngam P, Porter AR, Firdous S, Chiou J, Park MD, Hunt RL, Almufarriji FMF, Tan VY, Asiamah TK, McCausland JW, Fisher EL, Yeh AJ, Bae JS, Kobayashi SD, Wang JM, Barber DL, DeLeo FR, Otto M. Nguyen TH, et al. Nat Microbiol. 2022 Jan;7(1):62-72. doi: 10.1038/s41564-021-01012-9. Epub 2021 Dec 6. Nat Microbiol. 2022. PMID: 34873293 Free PMC article. - Complete genome sequence and annotation of the laboratory reference strain Shigella flexneri serotype 5a M90T and genome-wide transcriptional start site determination.
Cervantes-Rivera R, Tronnet S, Puhar A. Cervantes-Rivera R, et al. BMC Genomics. 2020 Apr 6;21(1):285. doi: 10.1186/s12864-020-6565-5. BMC Genomics. 2020. PMID: 32252626 Free PMC article.
References
- Mallick P, Schirle M, Chen SS, Flory MR, Lee H, et al. (2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol 25: 125–131. - PubMed
- Gallien S, Duriez E, Domon B (2011) Selected reaction monitoring applied to proteomics. J Mass Spect 46: 298–312. - PubMed
- Galan JE, Wolf-Watz H (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature 444: 567–573. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous