MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination - PubMed (original) (raw)
MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination
Mingyang Lu et al. Proc Natl Acad Sci U S A. 2013.
Abstract
Forward and backward transitions between epithelial and mesenchymal phenotypes play crucial roles in embryonic development and tissue repair. Aberrantly regulated transitions are also a hallmark of cancer metastasis. The genetic network that regulates these transitions appears to allow for the existence of a hybrid phenotype (epithelial/mesenchymal). Hybrid cells are endowed with mixed epithelial and mesenchymal characteristics, enabling specialized capabilities such as collective cell migration. Cell-fate determination between the three phenotypes is in fact regulated by a circuit composed of two highly interconnected chimeric modules--the miR-34/SNAIL and the miR-200/ZEB mutual-inhibition feedback circuits. Here, we used detailed modeling of microRNA-based regulation to study this core unit. More specifically, we investigated the functions of the two isolated modules and subsequently of the combined unit when the two modules are integrated into the full regulatory circuit. We found that miR-200/ZEB forms a tristable circuit that acts as a ternary switch, driven by miR-34/SNAIL, that is a monostable module that acts as a noise-buffering integrator of internal and external signals. We propose to associate the three stable states--(1,0), (high miR-200)/(low ZEB); (0,1), (low miR-200)/(high ZEB); and (1/2,1/2), (medium miR-200)/(medium ZEB)--with the epithelial, mesenchymal, and hybrid phenotypes, respectively. Our (1/2,1/2) state hypothesis is consistent with recent experimental studies (e.g., ZEB expression measurements in collectively migrating cells) and explains the lack of observed mesenchymal-to-hybrid transitions in metastatic cells and in induced pluripotent stem cells. Testable predictions of dynamic gene expression during complete and partial transitions are presented.
Keywords: cancer systems biology; metastable intermediate phenotypes; microRNA modeling; multistable decision circuit; partial EMT.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
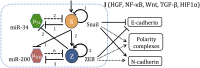
Fig. 1.
The regulatory network governing epithelial–mesenchymal plasticity. The unit is composed of two highly linked miR-TF chimeric circuits, the miR-34/SNAIL and the miR-200/ZEB mutually inhibiting loops. A solid arrow denotes transcriptional activation, and a solid bar denotes transcriptional inhibition. Dashed lines indicate microRNA-mediated translational regulation, and dotted lines represent indirect regulation. The numbers listed alongside each regulatory line represent the number of binding sites, as deduced from experiments.
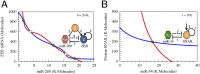
Fig. 2.
Dynamical system characteristics of the miR-200/ZEB and miR-34/SNAIL modules. The figures show the nullclines and the possible states in the phase-spaces corresponding to the two modules as are determined from Eqs. S4.2 and S4.3 in
SI Appendix
. (A) The miR-200/ZEB driven by signal SNAIL is tristable. Red nullcline is for dµ200/dt = 0 and dZ/dt = 0, and blue nullcline is for the condition dmZ/dt = 0 and dZ/dt = 0. (B) The miR-34/SNAIL is monostable. Blue nullcline is for the conditions dµ34/dt = 0 and dS/dt = 0, and red for dmS/dt = 0 and dS/dt = 0. The variables µ200, µ34, mZ, mS, Z, and S correspond to the levels of miR-200, miR-24, ZEB mRNA, SNAIL mRNA, ZEB, and SNAIL, respectively, as are defined in Eqs. S4.2 and S4.3. Green solid dots denote stable fixed points, and green unfilled circles denote unstable fixed points. The value of the SNAIL signal for A is taken from the stable fixed point in B. K Molecules, thousand molecules.
Fig. 3.
The microRNA-based circuit (MBC) modeling approach. (A) Schematic diagram for the MBC model. One or more microRNAs (µ) reversibly bind to mRNA (m) to form a µ–m complex. km and kµ are innate degradation rates of µ and m, respectively. µ can inhibit the translation of m [the translation rate is scaled by L(µ)] and degrade mRNA actively [at rate Ym(µ)]. The combined silencing effects can be characterized by P(µ). Also, µ can itself be degraded actively [at rate mYµ (µ) for µ microRNAs]. (B) The values of the L, Ym, Yµ as the functions of µ (scaled by the threshold µ0). The number of microRNA binding sites on the mRNA is taken to be six. A vertical dotted line is plotted to show the values at µ0. (C) The values of the shifted Hill function versus TF levels (scaled by the threshold TF0). Here, the Hill coefficient is three, and the fold change λ is 10.
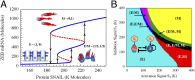
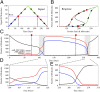
Fig. 4.
Bifurcation and phase-diagram of the driven miR-200/ZEB decision module. (A) Bifurcation of ZEB mRNA levels when driven by a signal S (
SI Appendix, Eqs. S4.2
) representing SNAIL. The bifurcation illustrates the possible coexistence (for some range of S) of three states: (i) the (1,0) state with high miR-200 and low ZEB, which corresponds to the epithelial (E) phenotype; (ii) the (0,1) state, which corresponds to the mesenchymal phenotype (M); (iii) the (1/2,1/2) state, which corresponds to the hybrid phenotype (E/M). Starting with the (1,0) state (E) and increasing SNAIL, the circuit undergoes a transition to the (1/2,1/2) state (E/M)—the first upward arrows on the right. Further increase in SNAIL leads to a transition from the (1/2,1/2) state to the (0,1) state (M)—the second upward arrows on the right. Starting from the (0,1) state and decreasing SNAIL yields a direct transition to the (1,0) state—the downward arrows further to the left. (B) The phase-diagram of the chimeric circuit when it is driven by two independent signals S1 and S2, as is illustrated in the Inset circuit. Each phase corresponds to a different combination of coexisting states. For example, in phase {E}, only state (1,0) is stable, and, in phase {E,E/M}, the states (1,0) and (1/2,1/2) can coexist.
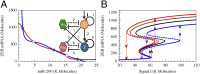
Fig. 5.
The phase-space and bifurcation diagram of the core regulatory unit. (A) The nullclines and the possible states in the phase-space corresponding to the combined regulatory unit, modeled by Eqs. S4.4 in
SI Appendix
. The blue nullcline is for the whole circuit when all ODEs are set to 0 except for mZ′ = 0 (red line), and the red nullcline is for the conditions when all ODEs are set to 0 except for µ200′ = 0. Green dots denote stable fixed points, and green unfilled circles denote unstable/saddle fixed points. (B) Bifurcation plots for the ZEB levels with respect to the driving signal I. Different lines show different cases of feedback from ZEB to miR-34 (regulation fold change is denoted by λZEB->miR34). Compared with the case without feedback (black, λZEB->miR34 = 1), strong repression (red, λZEB->miR34 = 0.2) or weaker repression (brown, λZEB->miR34 = 0.5) makes the bifurcation slightly shift to the left whereas activation (blue, λZEB->miR34 = 2.0) makes the bifurcation shift mainly to the right. The vertical dotted lines show the transitions among different states along the bifurcation curves.
Fig. 6.
Temporal dynamics of the complete and partial epithelial–hybrid–mesenchymal transition. (A) We simulated the effect of time-varying external signal. The basal signal level is shown in a purple dotted line. The input signal starts from zero (blue) at day 0 and increases to 100 K molecules (green) at day 25. Then, the input signal linearly decreases until day 50 (red). (B) The dynamics of ZEB in response to the input signal is shown in the SNAIL-ZEB phase-plane. The colors for the signal epoch and the corresponding response are matched. Both A and B also show arrows to clarify the time evolution. A bifurcation plot for ZEB with respect to SNAIL (black) is superimposed. (C) Temporal evolution of miR-200 (navy), ZEB mRNA (red), and ZEB protein (blue, scaled by 0.33 to fit in the plot). In A_–_C, we labeled the states at the basal signal with red dots. The plot shows that cells at the basal signal level are epithelial during EMT but are still mesenchymal during MET. Thus, the two-way transitions are dynamically asymmetric. The areas marked in gray are expanded in D to show that cells going from epithelial to mesenchymal pass through a partial EMT state with intermediate levels of miR-200 and ZEB during days 16–18, (E) but cells directly return from mesenchymal to epithelial without going through any intermediate state.
Similar articles
- Toward decoding the principles of cancer metastasis circuits.
Lu M, Jolly MK, Onuchic J, Ben-Jacob E. Lu M, et al. Cancer Res. 2014 Sep 1;74(17):4574-87. doi: 10.1158/0008-5472.CAN-13-3367. Cancer Res. 2014. PMID: 25183783 - A novel network integrating a miRNA-203/SNAI1 feedback loop which regulates epithelial to mesenchymal transition.
Moes M, Le Béchec A, Crespo I, Laurini C, Halavatyi A, Vetter G, Del Sol A, Friederich E. Moes M, et al. PLoS One. 2012;7(4):e35440. doi: 10.1371/journal.pone.0035440. Epub 2012 Apr 13. PLoS One. 2012. PMID: 22514743 Free PMC article. - An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition.
Gregory PA, Bracken CP, Smith E, Bert AG, Wright JA, Roslan S, Morris M, Wyatt L, Farshid G, Lim YY, Lindeman GJ, Shannon MF, Drew PA, Khew-Goodall Y, Goodall GJ. Gregory PA, et al. Mol Biol Cell. 2011 May 15;22(10):1686-98. doi: 10.1091/mbc.E11-02-0103. Epub 2011 Mar 16. Mol Biol Cell. 2011. PMID: 21411626 Free PMC article. - The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer?
Brabletz S, Brabletz T. Brabletz S, et al. EMBO Rep. 2010 Sep;11(9):670-7. doi: 10.1038/embor.2010.117. Epub 2010 Aug 13. EMBO Rep. 2010. PMID: 20706219 Free PMC article. Review. - ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer.
Hill L, Browne G, Tulchinsky E. Hill L, et al. Int J Cancer. 2013 Feb 15;132(4):745-54. doi: 10.1002/ijc.27708. Epub 2012 Jul 21. Int J Cancer. 2013. PMID: 22753312 Review.
Cited by
- Engaging plasticity: Differentiation therapy in solid tumors.
Bar-Hai N, Ishay-Ronen D. Bar-Hai N, et al. Front Pharmacol. 2022 Aug 11;13:944773. doi: 10.3389/fphar.2022.944773. eCollection 2022. Front Pharmacol. 2022. PMID: 36034865 Free PMC article. Review. - Epigenetic factor competition reshapes the EMT landscape.
Al-Radhawi MA, Tripathi S, Zhang Y, Sontag ED, Levine H. Al-Radhawi MA, et al. Proc Natl Acad Sci U S A. 2022 Oct 18;119(42):e2210844119. doi: 10.1073/pnas.2210844119. Epub 2022 Oct 10. Proc Natl Acad Sci U S A. 2022. PMID: 36215492 Free PMC article. - OVOL guides the epithelial-hybrid-mesenchymal transition.
Jia D, Jolly MK, Boareto M, Parsana P, Mooney SM, Pienta KJ, Levine H, Ben-Jacob E. Jia D, et al. Oncotarget. 2015 Jun 20;6(17):15436-48. doi: 10.18632/oncotarget.3623. Oncotarget. 2015. PMID: 25944618 Free PMC article. - Circulating Tumor Cells: From the Laboratory to the Cancer Clinic.
Sawabata N. Sawabata N. Cancers (Basel). 2020 Oct 20;12(10):3065. doi: 10.3390/cancers12103065. Cancers (Basel). 2020. PMID: 33092279 Free PMC article. - Cell plasticity in cancer cell populations.
Shen S, Clairambault J. Shen S, et al. F1000Res. 2020 Jun 22;9:F1000 Faculty Rev-635. doi: 10.12688/f1000research.24803.1. eCollection 2020. F1000Res. 2020. PMID: 32595946 Free PMC article. Review.
References
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. - PubMed
- Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10(7):445–457. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials