The landscape of candidate driver genes differs between male and female breast cancer - PubMed (original) (raw)
The landscape of candidate driver genes differs between male and female breast cancer
Ida Johansson et al. PLoS One. 2013.
Abstract
The rapidly growing collection of diverse genome-scale data from multiple tumor types sheds light on various aspects of the underlying tumor biology. With the objective to identify genes of importance for breast tumorigenesis in men and to enable comparisons with genes important for breast cancer development in women, we applied the computational framework COpy Number and EXpression In Cancer (CONEXIC) to detect candidate driver genes among all altered passenger genes. Unique to this approach is that each driver gene is associated with several gene modules that are believed to be altered by the driver. Thirty candidate drivers were found in the male breast cancers and 67 in the female breast cancers. We identified many known drivers of breast cancer and other types of cancer, in the female dataset (e.g. GATA3, CCNE1, GRB7, CDK4). In contrast, only three known cancer genes were found among male breast cancers; MAP2K4, LHP, and ZNF217. Many of the candidate drivers identified are known to be involved in processes associated with tumorigenesis, including proliferation, invasion and differentiation. One of the modules identified in male breast cancer was regulated by THY1, a gene involved in invasion and related to epithelial-mesenchymal transition. Furthermore, men with THY1 positive breast cancers had significantly inferior survival. THY1 may thus be a promising novel prognostic marker for male breast cancer. Another module identified among male breast cancers, regulated by SPAG5, was closely associated with proliferation. Our data indicate that male and female breast cancers display highly different landscapes of candidate driver genes, as only a few genes were found in common between the two. Consequently, the pathobiology of male breast cancer may differ from that of female breast cancer and can be associated with differences in prognosis; men diagnosed with breast cancer may consequently require different management and treatment strategies than women.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
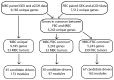
Figure 1. Flowchart outlining the steps in the CONEXIC analysis.
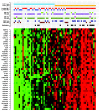
Figure 2. Heatmap of the genes in the module regulated by THY1 in MBC.
Red corresponds to up-regulation and green to down-regulation. The 66 MBC tumors are sorted according to their gene expression level of THY1.
Figure 3. Heatmap of the genes in the module regulated by SPAG5 in MBC.
Red corresponds to up-regulation and green to down-regulation. The 66 MBC tumors are sorted according to their gene expression level of SPAG5.
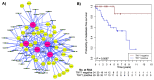
Figure 4. Graphic output of the LitVAn result for the MBC THY1 module and corresponding Kaplan-Meier survival analysis.
A) Significantly over-represented terms are represented as red circles and their association (graph edges) with multiple genes (yellow circles) from the module. The green dots represent the publication that significantly associates between the gene and the term, and the numbers in the green dots are the PubMed IDs for the respective publications. B) Distant metastasis free survival of the 66 MBC patients stratified by THY1 gene expression. The numbers below the plots indicate the number of patients at risk in each group at the given time points.
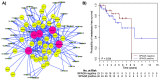
Figure 5. Graphic output of the LitVAn result for the MBC SPAG5 module and corresponding Kaplan-Meier survival analysis.
A) Significantly over-represented terms are represented as red circles and their association (graph edges) with multiple genes (yellow circles) from the module. The green dots represent the publication that significantly associates between the gene and the term, and the numbers in the green dots are the PubMed IDs for the respective publications. B) Distant metastasis free survival of the 66 MBC patients stratified by SPAG5 gene expression. The numbers below the plots indicate the number of patients at risk in each group at the given time points.
Similar articles
- An Entropy-Based Method for Identifying Mutual Exclusive Driver Genes in Cancer.
Song J, Peng W, Wang F. Song J, et al. IEEE/ACM Trans Comput Biol Bioinform. 2020 May-Jun;17(3):758-768. doi: 10.1109/TCBB.2019.2897931. Epub 2019 Feb 7. IEEE/ACM Trans Comput Biol Bioinform. 2020. PMID: 30763245 - Age-specific gene expression signatures for breast tumors and cross-species conserved potential cancer progression markers in young women.
Colak D, Nofal A, Albakheet A, Nirmal M, Jeprel H, Eldali A, Al-Tweigeri T, Tulbah A, Ajarim D, Malik OA, Inan MS, Kaya N, Park BH, Bin Amer SM. Colak D, et al. PLoS One. 2013 May 21;8(5):e63204. doi: 10.1371/journal.pone.0063204. Print 2013. PLoS One. 2013. PMID: 23704896 Free PMC article. - Inferring causal genomic alterations in breast cancer using gene expression data.
Tran LM, Zhang B, Zhang Z, Zhang C, Xie T, Lamb JR, Dai H, Schadt EE, Zhu J. Tran LM, et al. BMC Syst Biol. 2011 Aug 1;5:121. doi: 10.1186/1752-0509-5-121. BMC Syst Biol. 2011. PMID: 21806811 Free PMC article. - Characterization of potential driver mutations involved in human breast cancer by computational approaches.
Rajendran BK, Deng CX. Rajendran BK, et al. Oncotarget. 2017 Jul 25;8(30):50252-50272. doi: 10.18632/oncotarget.17225. Oncotarget. 2017. PMID: 28477017 Free PMC article. Review. - [Genetics and cancer: application to the breast].
Lidereau R, Nogues C. Lidereau R, et al. Arch Anat Cytol Pathol. 1995;43(1-2):5-11. Arch Anat Cytol Pathol. 1995. PMID: 7794027 Review. French.
Cited by
- Deciphering the molecular landscape: evolutionary progression from gynecomastia to aggressive male breast cancer.
Yang C, Wang Z, Qian L, Fu J, Sun H. Yang C, et al. Cell Oncol (Dordr). 2024 Oct;47(5):1831-1843. doi: 10.1007/s13402-024-00964-4. Epub 2024 Jun 18. Cell Oncol (Dordr). 2024. PMID: 38888848 - Demographics, Characteristics, and Outcomes of Male Breast Cancer Patients at the Methodist Health System, Dallas, USA.
Nong HQ, Eastwood D, Rodriguez K, Puri V. Nong HQ, et al. Cureus. 2023 Nov 25;15(11):e49394. doi: 10.7759/cureus.49394. eCollection 2023 Nov. Cureus. 2023. PMID: 38146568 Free PMC article. - Multigene Panel Sequencing Identifies a Novel Germline Mutation Profile in Male Breast Cancer Patients.
Al Saati A, Vande Perre P, Plenecassagnes J, Gilhodes J, Monselet N, Cabarrou B, Lignon N, Filleron T, Telly D, Perello-Lestrade E, Feillel V, Staub A, Martinez M, Chipoulet E, Collet G, Thomas F, Gladieff L, Toulas C. Al Saati A, et al. Int J Mol Sci. 2023 Sep 20;24(18):14348. doi: 10.3390/ijms241814348. Int J Mol Sci. 2023. PMID: 37762649 Free PMC article. - Single-Cell Transcriptional and Epigenetic Profiles of Male Breast Cancer Nominate Salient Cancer-Specific Enhancers.
Kim H, Wisniewska K, Regner MJ, Thennavan A, Spanheimer PM, Franco HL. Kim H, et al. Int J Mol Sci. 2023 Aug 22;24(17):13053. doi: 10.3390/ijms241713053. Int J Mol Sci. 2023. PMID: 37685859 Free PMC article. - Systemic Treatment of Breast Cancer. 1st Central-Eastern European Professional Consensus Statement on Breast Cancer.
Rubovszky G, Kocsis J, Boér K, Chilingirova N, Dank M, Kahán Z, Kaidarova D, Kövér E, Krakovská BV, Máhr K, Mriňáková B, Pikó B, Božović-Spasojević I, Horváth Z. Rubovszky G, et al. Pathol Oncol Res. 2022 Jul 11;28:1610383. doi: 10.3389/pore.2022.1610383. eCollection 2022. Pathol Oncol Res. 2022. PMID: 35898593 Free PMC article.
References
- Johansson I, Nilsson C, Berglund P, Strand C, Jönsson G et al. (2011) High-resolution genomic profiling of male breast cancer reveals differences hidden behind the similarities with female breast cancer. Breast Cancer Res Treat 129: 747–760. doi:10.1007/s10549-010-1262-8. PubMed: 21113657. - DOI - PubMed
- Johansson I, Nilsson C, Berglund P, Lauss M, Ringnér M et al. (2012) Gene expression profiling of primary male breast cancers reveals two unique subgroups and identifies N-acetyltransferase-1 (NAT1) as a novel prognostic biomarker. Breast Cancer Res 14: R31. doi:10.1186/bcr3116. PubMed: 22333393. - DOI - PMC - PubMed
- Tommasi S, Mangia A, Iannelli G, Chiarappa P, Rossi E et al. (2010) Gene copy number variation in male breast cancer by aCGH. Anal Cell Pathol (Amst) 33: 113–119. doi:10.1007/s13402-011-0041-9. PubMed: 2154757721045282. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This study was supported by grants from the Swedish Cancer Society, the G Nilsson Cancer Foundation, the Mrs. B Kamprad Foundation, the King Gustaf V’s Jubilee Foundation, the Lund University Hospital Research Foundation and governmental funding of clinical research within the national health services (ALF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous