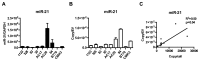MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development - PubMed (original) (raw)
. 2013 Oct 21;8(10):e78115.
doi: 10.1371/journal.pone.0078115. eCollection 2013.
Valya Ramakrishnan, Ryan Kim, Johan Skog, Ichiro Nakano, Sandeep Pingle, Juliya Kalinina, Wei Hua, Santosh Kesari, Ying Mao, Xandra O Breakefield, Fred H Hochberg, Erwin G Van Meir, Bob S Carter, Clark C Chen
Affiliations
- PMID: 24205116
- PMCID: PMC3804457
- DOI: 10.1371/journal.pone.0078115
MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development
Johnny C Akers et al. PLoS One. 2013.
Abstract
Glioblastoma cells secrete extra-cellular vesicles (EVs) containing microRNAs (miRNAs). Analysis of these EV miRNAs in the bio-fluids of afflicted patients represents a potential platform for biomarker development. However, the analytic algorithm for quantitative assessment of EV miRNA remains under-developed. Here, we demonstrate that the reference transcripts commonly used for quantitative PCR (including GAPDH, 18S rRNA, and hsa-miR-103) were unreliable for assessing EV miRNA. In this context, we quantitated EV miRNA in absolute terms and normalized this value to the input EV number. Using this method, we examined the abundance of miR-21, a highly over-expressed miRNA in glioblastomas, in EVs. In a panel of glioblastoma cell lines, the cellular levels of miR-21 correlated with EV miR-21 levels (p<0.05), suggesting that glioblastoma cells actively secrete EVs containing miR-21. Consistent with this hypothesis, the CSF EV miR-21 levels of glioblastoma patients (n=13) were, on average, ten-fold higher than levels in EVs isolated from the CSF of non-oncologic patients (n=13, p<0.001). Notably, none of the glioblastoma CSF harbored EV miR-21 level below 0.25 copies per EV in this cohort. Using this cut-off value, we were able to prospectively distinguish CSF derived from glioblastoma and non-oncologic patients in an independent cohort of twenty-nine patients (Sensitivity=87%; Specificity=93%; AUC=0.91, p<0.01). Our results suggest that CSF EV miRNA analysis of miR-21 may serve as a platform for glioblastoma biomarker development.
Conflict of interest statement
Competing Interests: We would like to declare that one of the co-author, Johan Skog, is an employee at Exosome Diagnostics. No other authors have any competing interests. This does not alter our adherence to all the PLOS ONE policies on sharing data and materials.
Figures
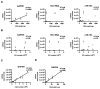
Figure 1. Relative abundance of GAPDH, 18S rRNA, and miR-103 in glioblastoma cell line derived EVs.
(A) GAPDH, 18S rRNA, and hsa-mir-103 levels in EVs isolated from 11 glioblastoma and 3 non-glioblastoma adherent cell lines, cultured under serum conditions. Transcript copy number was plotted against total RNA yield extracted from the EVs. (B) GAPDH, 18S rRNA, and hsa-mir-103 transcript copy number was plotted against the total number of EVs isolated for RNA extraction. (C) Cellular GAPDH transcript number tightly correlated with the number of cells collected for RNA extraction and (D) the amount of RNA recovered.
Figure 2. Relative abundance of GAPDH, 18S rRNA, and miR-103 in neurosphere glioblastoma line derived EVs.
(A) GAPDH, 18S rRNA, and hsa-mir-103 levels in EVs isolated from 9 glioblastoma neurophere lines, cultured under serum-free conditions. Transcript copy number was plotted against total RNA extracted from the EVs. (B) GAPDH, 18S rRNA, and hsa-mir-103 transcript copy number was plotted against the total number of EVs isolated for RNA extraction.
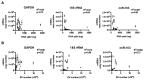
Figure 3. Relative abundance of GAPDH, 18S rRNA, and miR-103 in EV derived from CSF of glioblastoma and non-oncologic patients.
(A) GAPDH, 18S rRNA, and hsa-mir-103 levels in EVs isolated from the CSF of 13 glioblastoma and 14 non-oncologic patients. Transcript copy number was plotted against total RNA extracted from the EVs. (B) GAPDH, 18S rRNA, and hsa-mir-103 transcript copy number was plotted against the total number of EVs isolated for RNA extraction. The relative abundance of these transcripts in CSF EVs was approximately 10 fold lower than those of sera EV with abundance ranging between 1 transcript in 1 EV to 1 transcript in 105 EVs.
Figure 4. Detection of miR-21 in glioblastoma cell lines secreted EVs.
(A) The expression level of miR-21 was quantitatively assessed in nine neurosphere glioblastoma lines. (B) The abundance of miR-21 was assessed in EVs derived from nine neurosphere glioblastoma lines. (C) EV miR-21 levels correlate tightly with cellular miR-21 levels (R2 = 0.50, p<0.01).
Figure 5. Discrimination of glioblastoma disease status by CSF EV miR-21 analysis.
(A) Comparable levels of miR-21 level in sera EVs derived from 24 glioblastoma patients and 5 non-oncologic patients. (B) Elevated level of miR-21 in CSF EV derived from 13 glioblastoma patients relative to 14 non-oncologic patients. (C) miR-21 is detected in CSF EV but not in EV depleted CSF in samples isolated from five independent glioblastoma patients. Patient 1, 2, 3, 4, and 5 correspond to T2, T7, T10, T12, and T13 respectively from the UCSD glioblastoma cohort.
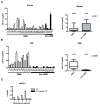
Figure 6. Elevated levels of CSF EV miR-21 are detected in glioblastoma patients.
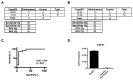
(A) Sensitivity, specificity, positive predictive, and negative predictive values associated with EV miR-21 level of <0.25 copy/EV as a discriminating threshold for glioblastoma disease status in the initial exploratory study. (B) Sensitivity, specificity, positive predictive, and negative predictive values associated with EV miR-21 level of <0.25 copy/EV as a discriminating threshold for glioblastoma disease status in a validation study. (C) Receiver Operating Characteristic Curve for EV miR-21 level of <0.25 copy/EV as a discriminating threshold for glioblastoma disease status. (D) Level of miR-21 in CSF EV from a glioblastoma patient at time of surgery and three months after a gross-total resection. The relative abundance of miR-21 in CSF EV was decreased by approximately 50-fold after surgical resection.
Similar articles
- miRNA contents of cerebrospinal fluid extracellular vesicles in glioblastoma patients.
Akers JC, Ramakrishnan V, Kim R, Phillips S, Kaimal V, Mao Y, Hua W, Yang I, Fu CC, Nolan J, Nakano I, Yang Y, Beaulieu M, Carter BS, Chen CC. Akers JC, et al. J Neurooncol. 2015 Jun;123(2):205-16. doi: 10.1007/s11060-015-1784-3. Epub 2015 Apr 23. J Neurooncol. 2015. PMID: 25903655 Free PMC article. - Extracellular vesicles derived from hypoxic glioma stem-like cells confer temozolomide resistance on glioblastoma by delivering miR-30b-3p.
Yin J, Ge X, Shi Z, Yu C, Lu C, Wei Y, Zeng A, Wang X, Yan W, Zhang J, You Y. Yin J, et al. Theranostics. 2021 Jan 1;11(4):1763-1779. doi: 10.7150/thno.47057. eCollection 2021. Theranostics. 2021. PMID: 33408780 Free PMC article. - Optimizing preservation of extracellular vesicular miRNAs derived from clinical cerebrospinal fluid.
Akers JC, Ramakrishnan V, Yang I, Hua W, Mao Y, Carter BS, Chen CC. Akers JC, et al. Cancer Biomark. 2016 Mar 25;17(2):125-32. doi: 10.3233/CBM-160609. Cancer Biomark. 2016. PMID: 27062568 Free PMC article. - The Roles of miRNA in Glioblastoma Tumor Cell Communication: Diplomatic and Aggressive Negotiations.
Buruiană A, Florian ȘI, Florian AI, Timiș TL, Mihu CM, Miclăuș M, Oșan S, Hrapșa I, Cataniciu RC, Farcaș M, Șușman S. Buruiană A, et al. Int J Mol Sci. 2020 Mar 12;21(6):1950. doi: 10.3390/ijms21061950. Int J Mol Sci. 2020. PMID: 32178454 Free PMC article. Review. - miRNA signature in glioblastoma: Potential biomarkers and therapeutic targets.
Rezaei O, Honarmand K, Nateghinia S, Taheri M, Ghafouri-Fard S. Rezaei O, et al. Exp Mol Pathol. 2020 Dec;117:104550. doi: 10.1016/j.yexmp.2020.104550. Epub 2020 Oct 1. Exp Mol Pathol. 2020. PMID: 33010295 Review.
Cited by
- EVmiRNA: a database of miRNA profiling in extracellular vesicles.
Liu T, Zhang Q, Zhang J, Li C, Miao YR, Lei Q, Li Q, Guo AY. Liu T, et al. Nucleic Acids Res. 2019 Jan 8;47(D1):D89-D93. doi: 10.1093/nar/gky985. Nucleic Acids Res. 2019. PMID: 30335161 Free PMC article. - Deep sequencing of circulating exosomal microRNA allows non-invasive glioblastoma diagnosis.
Ebrahimkhani S, Vafaee F, Hallal S, Wei H, Lee MYT, Young PE, Satgunaseelan L, Beadnall H, Barnett MH, Shivalingam B, Suter CM, Buckland ME, Kaufman KL. Ebrahimkhani S, et al. NPJ Precis Oncol. 2018 Dec 12;2:28. doi: 10.1038/s41698-018-0071-0. eCollection 2018. NPJ Precis Oncol. 2018. PMID: 30564636 Free PMC article. - The Study of Cerebrospinal Fluid microRNAs in Spinal Cord Injury and Neurodegenerative Diseases: Methodological Problems and Possible Solutions.
Baichurina I, Valiullin V, James V, Rizvanov A, Mukhamedshina Y. Baichurina I, et al. Int J Mol Sci. 2021 Dec 22;23(1):114. doi: 10.3390/ijms23010114. Int J Mol Sci. 2021. PMID: 35008540 Free PMC article. Review. - Microvesicles and Microvesicle-Associated microRNAs Reflect Glioblastoma Regression: Microvesicle-Associated miR-625-5p Has Biomarker Potential.
Simionescu N, Nemecz M, Petrovici AR, Nechifor IS, Buga RC, Dabija MG, Eva L, Georgescu A. Simionescu N, et al. Int J Mol Sci. 2022 Jul 29;23(15):8398. doi: 10.3390/ijms23158398. Int J Mol Sci. 2022. PMID: 35955533 Free PMC article. - Circulating biomarkers for gliomas.
Westphal M, Lamszus K. Westphal M, et al. Nat Rev Neurol. 2015 Oct;11(10):556-66. doi: 10.1038/nrneurol.2015.171. Epub 2015 Sep 15. Nat Rev Neurol. 2015. PMID: 26369507 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials