Human gut microbiome and risk for colorectal cancer - PubMed (original) (raw)
Human gut microbiome and risk for colorectal cancer
Jiyoung Ahn et al. J Natl Cancer Inst. 2013.
Abstract
We tested the hypothesis that an altered community of gut microbes is associated with risk of colorectal cancer (CRC) in a study of 47 CRC case subjects and 94 control subjects. 16S rRNA genes in fecal bacterial DNA were amplified by universal primers, sequenced by 454 FLX technology, and aligned for taxonomic classification to microbial genomes using the QIIME pipeline. Taxonomic differences were confirmed with quantitative polymerase chain reaction and adjusted for false discovery rate. All statistical tests were two-sided. From 794217 16S rRNA gene sequences, we found that CRC case subjects had decreased overall microbial community diversity (P = .02). In taxonomy-based analyses, lower relative abundance of Clostridia (68.6% vs 77.8%) and increased carriage of Fusobacterium (multivariable odds ratio [OR] = 4.11; 95% confidence interval [CI] = 1.62 to 10.47) and Porphyromonas (OR = 5.17; 95% CI = 1.75 to 15.25) were found in case subjects compared with control subjects. Because of the potentially modifiable nature of the gut bacteria, our findings may have implications for CRC prevention.
Figures
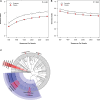
Figure 1.
Human gut microbiome in relation to colorectal cancer case-control status. A) Shannon diversity index in 47 colorectal cancer case subjects and 94 control subjects. B) Evenness index in 47 colorectal cancer case subjects and 94 control subjects. Rarefaction curves were estimated by bootstrapping of 500 random samples at 500 sequence increments. Alpha diversity (Shannon’s diversity and evenness indices) differences between case and control subjects were compared with t tests with Monte Carlo permutations using compare_alpha_diversity.py, a built-in function in the QIIME pipeline. C) Cladogram representation of gut microbiome taxa associated with colorectal cancer. Red indicates taxa enriched in colorectal cancer case subjects, and blue indicates taxa enriched in control subjects. Only taxa with nominal P less than .05 based on χ2 test (dichotomized) or Wilcoxon test (continuous) are labeled. The tests were two-sided. Figure was constructed using data presented in in Table 1.
Comment in
- Need for prospective cohort studies to establish human gut microbiome contributions to disease risk.
Mai V, Morris JG Jr. Mai V, et al. J Natl Cancer Inst. 2013 Dec 18;105(24):1850-1. doi: 10.1093/jnci/djt349. Epub 2013 Dec 6. J Natl Cancer Inst. 2013. PMID: 24316594 No abstract available.
Similar articles
- Re-purposing 16S rRNA gene sequence data from within case paired tumor biopsy and tumor-adjacent biopsy or fecal samples to identify microbial markers for colorectal cancer.
Shah MS, DeSantis T, Yamal JM, Weir T, Ryan EP, Cope JL, Hollister EB. Shah MS, et al. PLoS One. 2018 Nov 9;13(11):e0207002. doi: 10.1371/journal.pone.0207002. eCollection 2018. PLoS One. 2018. PMID: 30412600 Free PMC article. - Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations.
Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, Hayes RB, Goedert JJ. Sinha R, et al. PLoS One. 2016 Mar 25;11(3):e0152126. doi: 10.1371/journal.pone.0152126. eCollection 2016. PLoS One. 2016. PMID: 27015276 Free PMC article. - Combined Non-Invasive Prediction and New Biomarkers of Oral and Fecal Microbiota in Patients With Gastric and Colorectal Cancer.
Zhang C, Hu A, Li J, Zhang F, Zhong P, Li Y, Li Y. Zhang C, et al. Front Cell Infect Microbiol. 2022 May 19;12:830684. doi: 10.3389/fcimb.2022.830684. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35663463 Free PMC article. - Structural shift of gut microbiota during chemo-preventive effects of epigallocatechin gallate on colorectal carcinogenesis in mice.
Wang X, Ye T, Chen WJ, Lv Y, Hao Z, Chen J, Zhao JY, Wang HP, Cai YK. Wang X, et al. World J Gastroenterol. 2017 Dec 14;23(46):8128-8139. doi: 10.3748/wjg.v23.i46.8128. World J Gastroenterol. 2017. PMID: 29290650 Free PMC article. - Gut Microbial and Associated Metabolite Markers for Colorectal Cancer Diagnosis.
Alhhazmi AA, Alhamawi RM, Almisned RM, Almutairi HA, Jan AA, Kurdi SM, Almutawif YA, Mohammed-Saeid W. Alhhazmi AA, et al. Microorganisms. 2023 Aug 8;11(8):2037. doi: 10.3390/microorganisms11082037. Microorganisms. 2023. PMID: 37630597 Free PMC article. Review.
Cited by
- Racial and Ethnic Disparities in Colorectal Cancer Incidence Trends Across Regions of the United States From 2001 to 2020 - A United States Cancer Statistics Analysis.
Pankratz VS, Kanda D, Kosich M, Edwardson N, English K, Adsul P, Mishra SI. Pankratz VS, et al. Cancer Control. 2024 Jan-Dec;31:10732748241300653. doi: 10.1177/10732748241300653. Cancer Control. 2024. PMID: 39543981 Free PMC article. - Anemarrhena asphodeloides Bunge polysaccharides alleviate lipoteichoic acid-induced lung inflammation and modulate gut microbiota in mice.
Wen Y, Ullah H, Ma R, Farooqui NA, Li J, Alioui Y, Qiu J. Wen Y, et al. Heliyon. 2024 Oct 16;10(20):e39390. doi: 10.1016/j.heliyon.2024.e39390. eCollection 2024 Oct 30. Heliyon. 2024. PMID: 39469699 Free PMC article. - Parvimonas micra forms a distinct bacterial network with oral pathobionts in colorectal cancer patients.
Löwenmark T, Köhn L, Kellgren T, Rosenbaum W, Bronnec V, Löfgren-Burström A, Zingmark C, Larsson P, Dahlberg M, Schroeder BO, Wai SN, Ljuslinder I, Edin S, Palmqvist R. Löwenmark T, et al. J Transl Med. 2024 Oct 17;22(1):947. doi: 10.1186/s12967-024-05720-8. J Transl Med. 2024. PMID: 39420333 Free PMC article. - Water-soluble garlic polysaccharide (WSGP) improves ulcerative volitis by modulating the intestinal barrier and intestinal flora metabolites.
Shao X, Li J, Shao Q, Qu R, Ouyang X, Wang Y, Chen C. Shao X, et al. Sci Rep. 2024 Sep 14;14(1):21504. doi: 10.1038/s41598-024-72797-y. Sci Rep. 2024. PMID: 39277703 Free PMC article. - Correlation of gut microbial diversity to sight-threatening diabetic retinopathy.
Khan R, Sharma A, Ravikumar R, Sivaprasad S, Raman R. Khan R, et al. BMC Microbiol. 2024 Sep 13;24(1):342. doi: 10.1186/s12866-024-03496-x. BMC Microbiol. 2024. PMID: 39271995 Free PMC article.
References
- O’Keefe SJ. Nutrition and colonic health: the critical role of the microbiota. Curr Opin Gastroenterol. 2008;24(1):51–58 - PubMed
- Kinross JM, von Roon AC, Holmes E, Darzi A, Nicholson JK. The human gut microbiome: implications for future health care. Curr Gastroenterol Rep. 2008;10(4):396–403 - PubMed
- Horie H, Kanazawa K, Okada M, Narushima S, Itoh K, Terada A. Effects of intestinal bacteria on the development of colonic neoplasm: an experimental study. Eur J Cancer Prev. 1999;8(3):237–245 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA159036/CA/NCI NIH HHS/United States
- R03CA159414/CA/NCI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R03 CA159414/CA/NCI NIH HHS/United States
- R01CA159036/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical