Recruitment of PI4KIIIβ to coxsackievirus B3 replication organelles is independent of ACBD3, GBF1, and Arf1 - PubMed (original) (raw)
Recruitment of PI4KIIIβ to coxsackievirus B3 replication organelles is independent of ACBD3, GBF1, and Arf1
Cristina M Dorobantu et al. J Virol. 2014 Mar.
Abstract
Members of the Enterovirus (poliovirus [PV], coxsackieviruses, and human rhinoviruses) and Kobuvirus (Aichi virus) genera in the Picornaviridae family rely on PI4KIIIβ (phosphatidylinositol-4-kinase IIIβ) for efficient replication. The small membrane-anchored enteroviral protein 3A recruits PI4KIIIβ to replication organelles, yet the underlying mechanism has remained elusive. Recently, it was shown that kobuviruses recruit PI4KIIIβ through interaction with ACBD3 (acyl coenzyme A [acyl-CoA]-binding protein domain 3), a novel interaction partner of PI4KIIIβ. Therefore, we investigated a possible role for ACBD3 in recruiting PI4KIIIβ to enterovirus replication organelles. Although ACBD3 interacted directly with coxsackievirus B3 (CVB3) 3A, its depletion from cells by RNA interference did not affect PI4KIIIβ recruitment to replication organelles and did not impair CVB3 RNA replication. Enterovirus 3A was previously also proposed to recruit PI4KIIIβ via GBF1/Arf1, based on the known interaction of 3A with GBF1, an important regulator of secretory pathway transport and a guanine nucleotide exchange factor (GEF) of Arf1. However, our results demonstrate that inhibition of GBF1 or Arf1 either by pharmacological inhibition or depletion with small interfering RNA (siRNA) treatment did not affect the ability of 3A to recruit PI4KIIIβ. Furthermore, we show that a 3A mutant that no longer binds GBF1 was capable of recruiting PI4KIIIβ, even in ACBD3-depleted cells. Together, our findings indicate that unlike originally envisaged, coxsackievirus recruits PI4KIIIβ to replication organelles independently of ACBD3 and GBF1/Arf1.
Importance: A hallmark of enteroviral infection is the generation of new membranous structures to support viral RNA replication. The functionality of these "replication organelles" depends on the concerted actions of both viral nonstructural proteins and co-opted host factors. It is thus essential to understand how these structures are formed and which cellular components are key players in this process. GBF1/Arf1 and ACBD3 have been proposed to contribute to the recruitment of the essential lipid-modifying enzyme PI4KIIIβ to enterovirus replication organelles. Here we show that the enterovirus CVB3 recruits PI4KIIIβ by a mechanism independent of both GBF1/Arf1 and ACBD3. This study shows that the strategy employed by coxsackievirus to recruit PI4KIIIβ to replication organelles is far more complex than initially anticipated.
Figures
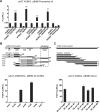
FIG 1
The GOLD domain of ACBD3 and aa 50 to 60 of CVB3 3A are crucial for interaction. (A) The ability of several picornavirus 3A proteins to interact with ACBD3 was determined using the mammalian two-hybrid system. Error bars represent standard deviations. Significant differences were calculated over the highest control, by paired t test. ***, P < 0.001. (B and C) Mapping of interaction domains of ACBD3 and CVB3 3A. Graphs at the bottom show interactions of CVB3 3A with truncated ACBD3 (B) and of ACBD3 with truncated CVB3 3A (C) in mammalian two-hybrid. Error bars represent standard deviations. Schematic representations of full-length and truncated variants of ACBD3 (B) and CVB3 3A (C) are depicted in the top portions.
FIG 2
ACBD3 directly interacts with CVB3 3A and PI4KIIIβ. (A) Rapid kinetic GST pulldown assay to study direct interaction between ACBD3-GST and CVB3 3A(1-60)-His, Strep-ACBD3, and PI4KIIIβ-GST or PI4KIIIβ-GST and 3A(1-60)-His. Unless otherwise specified, 2 μg of recombinant GST/GST-tagged protein was incubated for 2 h with Pierce glutathione magnetic beads and 2 μg of the other recombinant proteins. GST was used as a negative control. Following rapid washing, bound proteins were eluted off the beads and subjected to Western blot analysis. Proteins were detected with antibodies against the GST, Strep, and His tags. Purification of recombinant proteins is described in Materials and Methods. (B) Mammalian two-hybrid assay testing interactions between 3A, ACBD3, and PI4KIIIβ. Error bars represent standard deviations. Significant differences were calculated over the highest control, by paired t test. *, P < 0.1; **, P < 0.01; ***, P < 0.001.
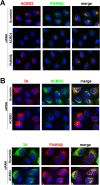
FIG 3
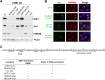
CVB3 recruits PI4KIIIβ to replication organelles independently of ACBD3. (A and B) HeLa R19 cells were reverse transfected with siRNA against PI4KIIIβ, ACBD3, or scramble siRNA as a control. At 48 h p.t., cells were mock infected (A) or infected with CVB3 wt at an MOI of 10. At 5 h p.i., cells were fixed and stained with antibodies against PI4KIIIβ and ACBD3 (A) or PI4KIIIβ, ACBD3, and CVB3 3A (B).
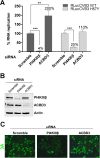
FIG 4
ACBD3 is not essential for CVB3 RNA replication. HeLa R19 cells were reverse transfected with siRNA against PI4KIIIβ or ACBD3 or scramble siRNA as a control. At 48 h p.t., cells were infected with CVB3-Rluc wt or CVB3-RLuc-3A H57Y at an MOI of 0.1 (A) or with CVB3 wt at an MOI of 10 (C). (A) At 8 h after infection with the luciferase-encoding viruses, cells were lysed and Renilla luciferase activity was determined as a measure of viral RNA replication. Error bars represent standard deviations. Significant differences were calculated by paired t test. **, P < 0.01; ***, P < 0.001. (B) Cells were harvested at 48 h p.t. and subjected to Western blot analysis to evaluate the knockdown efficiency. Actin was used as a loading control. (C) At 5 h after infection with CVB3 wt, cells were fixed and stained with antibodies against CVB3 3A.
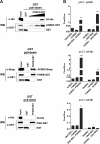
FIG 5
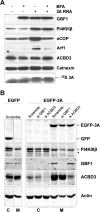
GBF1 is not essential for PI4KIIIβ recruitment by CVB3 3A. (A) In vitro HeLa S10 extract assay. RNA coding for CVB3 3A was translated in HeLa S10 cell extracts. Where indicated, the samples contained 80 μg/ml of BFA. Membranes were isolated by centrifugation and subjected to Western blot analysis to detect the membrane-associated proteins using antibodies against GBF1, PI4KIIIβ, αCOPI, and Arf1. Calnexin was used as a loading control. Efficiency of the translation reaction was assessed by [35S]methionine labeling. The arrow indicates specific signal for 3A (lowest blot). (B) Effect of GBF1 depletion on PI4KIIIβ recruitment to membranes by 3A. HeLa R19 cells were reverse transfected with siRNA against GBF1 or ACBD3 or scramble siRNA as a control. At 48 h p.t., cells were transfected with CVB3 3A-EGFP or GFP as a control. At 24 h after DNA transfection, cells were harvested, resuspended in PBS to separate membranes (M) from cytosol (C) by three freeze-thaw cycles, and subjected to Western blot analysis using antibodies against GBF1, ACBD3, PI4KIIIβ, and GFP. Actin was used as a loading control. The star indicates a nonspecific band.
FIG 6
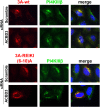
CVB3 3A mutants recruit PI4KIIIβ in the absence of GBF1. (A) In vitro translation assay. RNA coding for wt or mutant CVB3 3A was translated in HeLa S10 cell extracts. Isolated membranes were analyzed by Western blotting to detect the membrane-associated proteins, using antibodies against GBF1, Arf1, and PI4KIIIβ. Efficiency of the translation reaction was assessed by [35S]methionine labeling (the faint signal in the H2O sample corresponds to the [35S]methionine front). (B) To study PI4KIIIβ recruitment in intact cells, HeLa R19 cells were transfected with myc-tagged CVB3 3A-wt or the indicated mutants. The next day, cells were fixed and stained with antibodies against the myc tag or endogenous PI4KIIIβ. Shown are representative immunofluorescence example images of proteins listed in the table in panel C with recruitment of PI4KIIIβ by different 3A proteins. (C) CVB3 3A amino acid sequence. Residues replaced by alanine are indicated by stars. The Ser insertion at position 16 and the C-terminal hydrophobic domain are also depicted. The table outlines the ability of the different 3A mutants to interact with GBF1 in the mammalian two-hybrid system (30) and to recruit PI4KIIIβ.
FIG 7
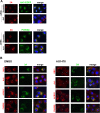
Arf1 is dispensable for PI4KIIIβ recruitment by CVB3 3A. (A) Effect of Arf1 depletion on PI4KIIIβ recruitment to membranes by 3A. HeLa R19 cells were reverse transfected with siRNA against Arf1 or scramble siRNA as a control. At 48 h posttransfection, cells were transfected with Arf1-EGFP and CVB3 3A. At 24 h after DNA transfection, cells were fixed and stained with antibodies against 3A and PI4KIIIβ. In cells treated with scramble siRNA, the degree of 3A and Arf1-EGFP coexpression was greater than 90%. (B) BGM cells were infected with CVB3 wt at an MOI of 10, followed by treatment with DMSO or 25 μM AG1478. At 5 h p.i., cells were stained with antibodies against CVB3 3A and endogenous GBF1, Arf1, ACBD3, or PI4KIIIβ. Uninfected cells are marked by asterisks.
FIG 8
GBF1, Arf1, and ACBD3 are simultaneously dispensable for PI4KIIIβ recruitment by CVB3 3A. HeLa R19 cells were reverse transfected with scramble siRNA or siRNA against ACBD3. At 48 h p.t., cells were transfected with myc-tagged CVB3 3A-wt or the REIKI(6-10)A mutant. At 24 h p.t., cells were fixed and stained with antibodies against PI4KIIIβ or the myc tag to detect 3A.
Similar articles
- ACBD3 Is an Essential Pan-enterovirus Host Factor That Mediates the Interaction between Viral 3A Protein and Cellular Protein PI4KB.
Lyoo H, van der Schaar HM, Dorobantu CM, Rabouw HH, Strating JRPM, van Kuppeveld FJM. Lyoo H, et al. mBio. 2019 Feb 12;10(1):e02742-18. doi: 10.1128/mBio.02742-18. mBio. 2019. PMID: 30755512 Free PMC article. - GBF1- and ACBD3-independent recruitment of PI4KIIIβ to replication sites by rhinovirus 3A proteins.
Dorobantu CM, Ford-Siltz LA, Sittig SP, Lanke KH, Belov GA, van Kuppeveld FJ, van der Schaar HM. Dorobantu CM, et al. J Virol. 2015 Feb;89(3):1913-8. doi: 10.1128/JVI.02830-14. Epub 2014 Nov 19. J Virol. 2015. PMID: 25410869 Free PMC article. - The 3A protein from multiple picornaviruses utilizes the golgi adaptor protein ACBD3 to recruit PI4KIIIβ.
Greninger AL, Knudsen GM, Betegon M, Burlingame AL, Derisi JL. Greninger AL, et al. J Virol. 2012 Apr;86(7):3605-16. doi: 10.1128/JVI.06778-11. Epub 2012 Jan 18. J Virol. 2012. PMID: 22258260 Free PMC article. - Emerging Role for Acyl-CoA Binding Domain Containing 3 at Membrane Contact Sites During Viral Infection.
Lu Y, Song S, Zhang L. Lu Y, et al. Front Microbiol. 2020 Apr 8;11:608. doi: 10.3389/fmicb.2020.00608. eCollection 2020. Front Microbiol. 2020. PMID: 32322249 Free PMC article. Review. - The role of phosphatidylinositol 4-kinases and phosphatidylinositol 4-phosphate during viral replication.
Delang L, Paeshuyse J, Neyts J. Delang L, et al. Biochem Pharmacol. 2012 Dec 1;84(11):1400-8. doi: 10.1016/j.bcp.2012.07.034. Epub 2012 Aug 8. Biochem Pharmacol. 2012. PMID: 22885339 Free PMC article. Review.
Cited by
- An efficient trans complementation system for in vivo replication of defective poliovirus mutants.
Arita M. Arita M. J Virol. 2024 Jul 23;98(7):e0052324. doi: 10.1128/jvi.00523-24. Epub 2024 Jun 5. J Virol. 2024. PMID: 38837378 Free PMC article. - Curcumol inhibits EMCV replication by activating CH25H and inhibiting the formation of ROs.
Zheng J, Sun P, Sun N, Hao Z, Fan K, Yin W, Khan A, Guo J, Zheng X, Li H. Zheng J, et al. BMC Vet Res. 2022 Dec 26;18(1):453. doi: 10.1186/s12917-022-03531-x. BMC Vet Res. 2022. PMID: 36572890 Free PMC article. - A plant virus hijacks phosphatidylinositol-3,5-bisphosphate to escape autophagic degradation in its insect vector.
Wang H, Zhang J, Liu H, Wang M, Dong Y, Zhou Y, Wong SM, Xu K, Xu Q. Wang H, et al. Autophagy. 2023 Apr;19(4):1128-1143. doi: 10.1080/15548627.2022.2116676. Epub 2022 Sep 10. Autophagy. 2023. PMID: 36093594 Free PMC article. - Defining the proximal interaction networks of Arf GTPases reveals a mechanism for the regulation of PLD1 and PI4KB.
Li FL, Wu Z, Gao YQ, Bowling FZ, Franklin JM, Hu C, Suhandynata RT, Frohman MA, Airola MV, Zhou H, Guan KL. Li FL, et al. EMBO J. 2022 Sep 1;41(17):e110698. doi: 10.15252/embj.2022110698. Epub 2022 Jul 17. EMBO J. 2022. PMID: 35844135 Free PMC article. - CRISPR-Cas9-Based Technology for Studying Enteric Virus Infection.
Hirano J, Murakami K, Hayashi T. Hirano J, et al. Front Genome Ed. 2022 Jun 8;4:888878. doi: 10.3389/fgeed.2022.888878. eCollection 2022. Front Genome Ed. 2022. PMID: 35755450 Free PMC article. Review.
References
- Romero-Brey I, Merz A, Chiramel A, Lee J-Y, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. 2012. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 8:e1003056. 10.1371/journal.ppat.1003056 - DOI - PMC - PubMed
- Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CKE, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. 10.1016/j.chom.2009.03.007 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources