Dynamic Epstein-Barr virus gene expression on the path to B-cell transformation - PubMed (original) (raw)
Review
Dynamic Epstein-Barr virus gene expression on the path to B-cell transformation
Alexander M Price et al. Adv Virus Res. 2014.
Abstract
Epstein-Barr virus (EBV) is an oncogenic human herpesvirus in the γ-herpesvirinae subfamily that contains a 170-180kb double-stranded DNA genome. In vivo, EBV commonly infects B and epithelial cells and persists for the life of the host in a latent state in the memory B-cell compartment of the peripheral blood. EBV can be reactivated from its latent state, leading to increased expression of lytic genes that primarily encode for enzymes necessary to replicate the viral genome and structural components of the virion. Lytic cycle proteins also aid in immune evasion, inhibition of apoptosis, and the modulation of other host responses to infection. In vitro, EBV has the potential to infect primary human B cells and induce cellular proliferation to yield effectively immortalized lymphoblastoid cell lines, or LCLs. EBV immortalization of B cells in vitro serves as a model system for studying EBV-mediated lymphomagenesis. While much is known about the steady-state viral gene expression within EBV-immortalized LCLs and other EBV-positive cell lines, relatively little is known about the early events after primary B-cell infection. It was previously thought that upon latent infection, EBV only expressed the well-characterized latency-associated transcripts found in LCLs. However, recent work has characterized the early, but transient, expression of lytic genes necessary for efficient transformation and delayed responses in the known latency genes. This chapter summarizes these recent findings that show how dynamic and controlled expression of multiple EBV genes can control the activation of B cells, entry into the cell cycle, the inhibition of apoptosis, and innate and adaptive immune responses.
Keywords: EBNA; Epstein–Barr virus; Herpesvirus; LMP; Latency; Lytic; Viral gene expression; Viral transformation.
© 2014 Elsevier Inc. All rights reserved.
Figures
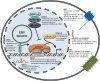
Figure 1. Latency III gene expression in a Lymphoblastoid Cell Line
Schematic diagram of latency proteins and RNAs expressed at steady-state in EBV-transformed LCLs. The nucleus is depicted by the inner, gray-shaded dotted circle. The latent membrane proteins (LMPs) are depicted in the plasma membrane as monomers, but likely exist as multimers and signal from multiple cellular membranes. The EBNA proteins are all shown as nuclear, but may have functions in the cytoplasm as well (e.g. EBNA-LP).
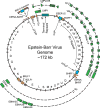
Figure 2. Schematic of the Epstein-Barr Virus genome
Letters on the inner edge of the circular genome denote BamHI digestion fragments. Cis-acting elements within the genome, such as the origin of plasmid replication (oriP), the two origins of lytic replication (oriLyt) and the terminal repeats formed when the linear genome is circularized are denoted in blue squares. Lytic genes that appear to be active early after infection in the pre-latent phase are shown in orange boxes. Coding exons for the latency genes are shown in green boxes. EBV latent mRNAs can be initiated from different promoters depending on latency type and time after infection: the W promoter (Wp), the C Promoter (Cp), the Q promoter (Qp, only in Latency I), and the LMP promoters are labeled. The unspliced pre-mRNAs driven from these promoters is shown as a dotted line. EBV encoded noncoding RNAs, such as the miR-BHRF1 cluster, the miR-BART cluster, and the EBERs are shown as red triangles.
Figure 3. EBV latency mRNAs are expressed as alternative isoforms and distinct transcripts
At the top is a schematized EBV linear genome showing the positions of latency gene exons in black boxes and BamHI fragment names listed below (not to scale). Also shown on the genome are the terminal repeats (open boxes), the C promoter (Cp, green boxes), the W promoter (Wp, yellow box), the Q promoter (Qp, blue box), the bi-directional Latent Membrane Protein promoter (LMPp, purple boxes), the LMP2A-specific promoter (purple box), and the canonical EBNA poly-adenylation sites (pA, arrow). The ORF-containing exon of the lytic gene BHRF1 is shown as an orange box. All coding exons are shown as full height boxes while non-coding exons are half-height. Early after infection latency transcripts are initiated primarily from the W promoter, as shown in yellow. The special instance of alternative splicing between the upstream Wp or Cp splice donor and the W1 or W1′ exon that leads to inclusion of an ATG start codon and EBNA-LP protein production is shown (Inset). After EBNA2 and EBNA-LP production reach a significant level early after infection, the C promoter is activated and transcribes the rest of the EBNAs, as shown in green. Later in infection, the LMP promoters are active and LMP1, LMP2A, and LMP2B and transcribed and spliced as shown in purple. In Latency I only the Q promoter is active to transcribe EBNA1.
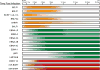
Figure 4. Timing of latent and lytic gene expression after infection by EBV
Relative expression levels of the RNA species are shown as shaded bars. Dark shading is indicative of the relative maximum amount of expression of the given RNA over the course of B cell growth transformation by EBV. Lytic genes expressed during the pre-latent phase are shown in orange, latency genes are shown in green, and noncoding RNAs are shown in red.
Similar articles
- RNA Sequencing Analyses of Gene Expression during Epstein-Barr Virus Infection of Primary B Lymphocytes.
Wang C, Li D, Zhang L, Jiang S, Liang J, Narita Y, Hou I, Zhong Q, Zheng Z, Xiao H, Gewurz BE, Teng M, Zhao B. Wang C, et al. J Virol. 2019 Jun 14;93(13):e00226-19. doi: 10.1128/JVI.00226-19. Print 2019 Jul 1. J Virol. 2019. PMID: 31019051 Free PMC article. - Human B cells on their route to latent infection--early but transient expression of lytic genes of Epstein-Barr virus.
Kalla M, Hammerschmidt W. Kalla M, et al. Eur J Cell Biol. 2012 Jan;91(1):65-9. doi: 10.1016/j.ejcb.2011.01.014. Epub 2011 Mar 29. Eur J Cell Biol. 2012. PMID: 21450364 Review. - Regulation and dysregulation of Epstein-Barr virus latency: implications for the development of autoimmune diseases.
Niller HH, Wolf H, Minarovits J. Niller HH, et al. Autoimmunity. 2008 May;41(4):298-328. doi: 10.1080/08916930802024772. Autoimmunity. 2008. PMID: 18432410 Review. - The Epstein-Barr virus BamHI C promoter is not essential for B cell immortalization in vitro, but it greatly enhances B cell growth transformation.
Tierney RJ, Nagra J, Rowe M, Bell AI, Rickinson AB. Tierney RJ, et al. J Virol. 2015 Mar;89(5):2483-93. doi: 10.1128/JVI.03300-14. Epub 2014 Dec 24. J Virol. 2015. PMID: 25540367 Free PMC article. - Maintenance of Epstein-Barr Virus Latent Status by a Novel Mechanism, Latent Membrane Protein 1-Induced Interleukin-32, via the Protein Kinase Cδ Pathway.
Lai KY, Chou YC, Lin JH, Liu Y, Lin KM, Doong SL, Chen MR, Yeh TH, Lin SJ, Tsai CH. Lai KY, et al. J Virol. 2015 Jun;89(11):5968-80. doi: 10.1128/JVI.00168-15. Epub 2015 Mar 25. J Virol. 2015. PMID: 25810549 Free PMC article.
Cited by
- Epigenetic Mechanisms in Latent Epstein-Barr Virus Infection and Associated Cancers.
Torne AS, Robertson ES. Torne AS, et al. Cancers (Basel). 2024 Feb 29;16(5):991. doi: 10.3390/cancers16050991. Cancers (Basel). 2024. PMID: 38473352 Free PMC article. Review. - Epstein-Barr Virus and Systemic Autoimmune Diseases.
Houen G, Trier NH. Houen G, et al. Front Immunol. 2021 Jan 7;11:587380. doi: 10.3389/fimmu.2020.587380. eCollection 2020. Front Immunol. 2021. PMID: 33488588 Free PMC article. Review. - Identification of the xenograft and its ascendant sphere-forming cell line as belonging to EBV-induced lymphoma, and characterization of the status of sphere-forming cells.
Dolgova EV, Petrova DD, Proskurina AS, Ritter GS, Kisaretova PE, Potter EA, Efremov YR, Bayborodin SI, Karamysheva TV, Romanenko MV, Netesov SV, Taranov OS, Ostanin AA, Chernykh ER, Bogachev SS. Dolgova EV, et al. Cancer Cell Int. 2019 May 6;19:120. doi: 10.1186/s12935-019-0842-x. eCollection 2019. Cancer Cell Int. 2019. PMID: 31080361 Free PMC article. - Epstein-Barr virus as a potentiator of autoimmune diseases.
Robinson WH, Younis S, Love ZZ, Steinman L, Lanz TV. Robinson WH, et al. Nat Rev Rheumatol. 2024 Nov;20(11):729-740. doi: 10.1038/s41584-024-01167-9. Epub 2024 Oct 10. Nat Rev Rheumatol. 2024. PMID: 39390260 Review. - Molecular mechanisms of viral oncogenesis in humans.
Krump NA, You J. Krump NA, et al. Nat Rev Microbiol. 2018 Nov;16(11):684-698. doi: 10.1038/s41579-018-0064-6. Nat Rev Microbiol. 2018. PMID: 30143749 Free PMC article. Review.
References
- Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181:595–608. - PubMed
- Allday MJ, Crawford DH, Griffin BE. Epstein-Barr virus latent gene expression during the initiation of B cell immortalization. J Gen Virol. 1989;70(Pt 7):1755–1764. - PubMed
Publication types
MeSH terms
Grants and funding
- 5T32CA009111/CA/NCI NIH HHS/United States
- R01 CA140337/CA/NCI NIH HHS/United States
- 5P30 AI064518/AI/NIAID NIH HHS/United States
- F31 CA180451/CA/NCI NIH HHS/United States
- T32 CA009111/CA/NCI NIH HHS/United States
- 1R01-CA140337/CA/NCI NIH HHS/United States
- P30 AI064518/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources