The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein - PubMed (original) (raw)
Hyun-Woo Suh 2, Young Ho Jeon 3, Eunha Hwang 4, Loi T Nguyen 5, Jeonghun Yeom 6, Seung-Goo Lee 7, Cheolju Lee 6, Kyung Jin Kim 8, Beom Sik Kang 8, Jin-Ok Jeong 9, Tae-Kwang Oh 5, Inpyo Choi 2, Jie-Oh Lee 10, Myung Hee Kim 11
Affiliations
- PMID: 24389582
- PMCID: PMC3941024
- DOI: 10.1038/ncomms3958
Free PMC article
The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein
Jungwon Hwang et al. Nat Commun. 2014.
Free PMC article
Abstract
The redox-dependent inhibition of thioredoxin (TRX) by thioredoxin-interacting protein (TXNIP) plays a pivotal role in various cancers and metabolic syndromes. However, the molecular mechanism of this regulation is largely unknown. Here, we present the crystal structure of the TRX-TXNIP complex and demonstrate that the inhibition of TRX by TXNIP is mediated by an intermolecular disulphide interaction resulting from a novel disulphide bond-switching mechanism. Upon binding to TRX, TXNIP undergoes a structural rearrangement that involves switching of a head-to-tail interprotomer Cys63-Cys247 disulphide between TXNIP molecules to an interdomain Cys63-Cys190 disulphide, and the formation of a de novo intermolecular TXNIP Cys247-TRX Cys32 disulphide. This disulphide-switching event unexpectedly results in a domain arrangement of TXNIP that is entirely different from those of other arrestin family proteins. We further show that the intermolecular disulphide bond between TRX and TXNIP dissociates in the presence of high concentrations of reactive oxygen species. This study provides insight into TRX and TXNIP-dependent cellular regulation.
Figures
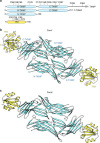
Figure 1. Overall structure of the heterodimeric complex of TRX and TXNIP in the asymmetric unit.
(a) Schematic representation of the TXNIP (cyan) and TRX (yellow) constructs used in this study, showing the locations of the cysteines. (b,c) Ribbon representations of the structures of the TRX(C35A)–T–TXNIP(C120S/C170S/C205S/C267S) complex (referred to as Com1) (b) and the TRX(C35A)–T–TXNIP(C170S/C205S/C267S) complex (referred to as Com2) (c). The structures of Com1 and Com2 were determined at resolutions of 2.0 Å and 2.7 Å, respectively. There are two heterodimeric complexes of TRX and TXNIP in the asymmetric unit. The β-sheets and disordered regions of TXNIP are shown in cyan and by white dashed lines, respectively. The α-helices and β-sheets of TRX are shown in yellow. The N-terminal TXNIP (N-TXNIP) and C-terminal TXNIP (C-TXNIP) domains are indicated.
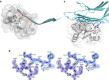
Figure 2. Representative structure of the TRX and TXNIP complex and structural comparison between TXNIP and arrestin.
(a) TRX interacts exclusively with C-TXNIP in the complex structure. The β-strands and disordered regions of TXNIP are shown in cyan and as grey dashed lines, respectively. The cysteines in TXNIP are displayed as white carbon atoms. Cys63 is located 39 Å from Cys247 and 14.1 Å from Cys120. (b) TRX-bound TXNIP (cyan) superimposed onto mVPS26B (magenta) based on the C-terminal domains. The disordered regions of TXNIP and mVPS26B are shown as dashed cyan and magenta lines, respectively. (c) Superimposition of N-TXNIP and C-TXNIP (cyan) onto N-mVPS26B and C-mVPS26B (magenta). The disordered region of C-TXNIP is shown as a dashed white line. (d) The electrostatic surface potential of TXNIP shows a highly basic C-terminal domain and a relatively negative electrostatic N-terminal domain.
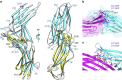
Figure 3. Detailed characterization of the TRX–TXNIP interaction.
(a) The critical β18 strand (cyan) of TXNIP that interacts with TRX is shown. The cleft formed by residues in the active site of TRX is highlighted in pink. The TRX Cys32 and TXNIP Cys247 residues that form an intermolecular disulphide bond are shown as white and cyan carbon atoms, respectively. (b) Stereo image of the 2Fo-Fc electron density map showing the intermolecular disulphide bond between the TRX Cys32 (white) and TXNIP Cys247 (cyan) residues contoured at 1.5σ. (c) Detailed depiction of the interactions between TRX (white) and TXNIP (cyan). The backbone–backbone interactions between TRX Met74 and TXNIP Cys247, and TRX Ala92 and TXNIP Gly245, are displayed as dashed red lines. The salt bridge between TRX Asp60 and TXNIP Arg251 and the hydrogen bond between TRX Gly33 and TXNIP Glu202 are shown as dashed black lines. Strands β15 and β18 in TXNIP are also indicated.
Figure 4. TXNIP molecules form interprotomer disulphide bonds between Cys63 and Cys247.
(a,b) Co-immunoprecipitation assays were performed using lysates from 293 T cells transfected with combinations of FLAG-tagged (F), HA-tagged, and GST-tagged full-length TXNIP, T–TXNIP, or mutant TXNIP. Immobilized proteins on FLAG-agarose beads or glutathione beads were visualized by immunoblotting using anti-HA, anti-FLAG, or anti-GST antibodies. One percent of the WCL was used as the input. (c) Proteomic analysis of the interprotomer-interacting TXNIP molecules fractionated by SDS–PAGE under non-reducing conditions. The MS/MS spectrum shows the interprotomer disulphide bond between Cys63 and Cys247 identified as 54VLWMQGSQQ
C
K64-240GNHISGT
C
ASWR251. Doubly charged [M+2H]+ peptide ions at m/z 1353.64 were fragmented via higher-energy collisional dissociation. Matched peaks are shown in red. The ion types of matched peaks are written in red for b- and y-ions and blue for ions from C-S and S-S bond cleavages. Annotations used are: P, strand VLWMQGSQQCK; p, strand GNHISGTCASWR; B and Y, ions from P; b and y, ions from p; p+32, persulfide ion of p formed by C-S bond cleavage reactions. (d) The effect of TRX on the interaction between TXNIP molecules. Co-immunoprecipitation assays were performed using lysates from 293 T cells transfected with FLAG-tagged TXNIP, HA-tagged TXNIP and FLAG-tagged TRX. Immobilized proteins on HA-agarose beads were visualized by immunoblotting using anti-HA and anti-FLAG antibodies. (a,b,d) The results are representative of at least two independent experiments with similar results.
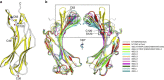
Figure 5. N-TXNIP undergoes a Cys63-mediated conformational change.
(a) Ribbon representation of N-TXNIP showing the intramolecular disulphide bond between Cys63 and Cys120. The locations of other cysteine residues are also shown. (b) N-TXNIP structures were superimposed onto the N-TXNIP(K5A/K6A) structure (grey). The structure of N-TXNIP(C36S/C49S/K64A/C120S) (Protein Data Bank accession code 4GEI) is shown in yellow. The structures of the crystallographically independent molecules of N-TXNIP(C36S/C49S/C120S) (Protein Data Bank accession code 4GEJ) are shown in different colours. The N-terminal domain of Com1 (red) has a similar conformation to the D-chain structure (wheat) of N-TXNIP(C36S/C49S/C120S). The region that undergoes significant structural changes between the N-TXNIP structures is indicated by the black dashed line box. The locations of the Cys63, Cys120 and Ser120 residues are indicated.
Figure 6. The interaction between TRX and TXNIP involves disulphide bond switching.
(a) TXNIP undergoes disulphide bond switching via a significant Cys63-mediated conformational change. The N-TXNIP structure (yellow) is superimposed onto the T–TXNIP structure (cyan). The interdomain disulphide bond between Cys63 and Cys190 in T–TXNIP and the intramolecular disulphide bond between Cys63 and Cys120 in N-TXNIP are indicated. (b) The interdomain disulphide bond formed between Cys63 of N-TXNIP (magenta) and Cys190 of C-TXNIP (cyan) is located at the centre of the interdomain interface. The residues involved in the interface are depicted using stick representations. The β5, β6, and β14 strands are also indicated. (c) A side-to-side interaction occurs between the TXNIP domains. The interdomain interactions between the N-terminal strand β6 (magenta) and the C-terminal strand β19 (cyan) are shown.
Figure 7. ROS directly affect the intermolecular disulphide bond between TRX and TXNIP.
(a,b) Size-exclusion chromatography analyses of the effect of ROS on the interaction between TRX and TXNIP. The TRX-T–TXNIP(C120S/C170S/C205S/C267S) complex was incubated with 0, 5, or 10 mM H2O2 at 37 °C for 30 min (a) and the TRX(C73A)–T–TXNIP(C36S/C49S/C120S/C170S/C205S/C267S) complex was incubated with 0 or 3 mM diamide at 37 °C for 30 min (b). The reaction products were injected onto a HiLoad 16/60 Superdex 75 gel filtration column at room temperature. (c) The TRX(C35A)–T–TXNIP(C120S/C170S/C205S/C267S) complex was incubated with 3.3 mM diamide at 37 °C for 60 min. The reaction products were injected into a Superdex 75 10/300 GL gel filtration column at room temperature. (a–c) All eluted proteins were analysed by SDS–PAGE. T–TXNIP(m), T–TXNIP mutant; LS, loading solution.
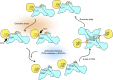
Figure 8. Proposed molecular mechanism of the negative regulation of TRX by TXNIP.
The inhibition of TRX by TXNIP is regulated by a redox-dependent disulphide bond-switching mechanism. Under normoxic conditions, TXNIP binds to TRX through an intermolecular disulphide bond formed between TXNIP Cys247 and TRX Cys32 and inhibits its reducing activity. During oxidative stress, high levels of ROS may cause further oxidation of this bond and lead to the dissociation of TXNIP from TRX. ROS also likely trigger antioxidant pathways to restore a reducing environment. After restoration of a reductive redox potential, the active cysteines of TRX may interact with the interprotomer disulphide bond between Cys63 and Cys247 of TXNIP molecules. Subsequently, TXNIP molecules would undergo a structural rearrangement that involves switching of the interprotomer disulphide bond between TXNIP molecules to an interdomain Cys63-Cys190 disulphide bond, and the formation of a de novo intermolecular TXNIP Cys247-TRX Cys32 disulphide bond. See discussion for a full description. TRX and TXNIP are shown in yellow and cyan, respectively. Cysteine residues forming disulphide bonds are displayed in brown.
Similar articles
- The interaction of thioredoxin with Txnip. Evidence for formation of a mixed disulfide by disulfide exchange.
Patwari P, Higgins LJ, Chutkow WA, Yoshioka J, Lee RT. Patwari P, et al. J Biol Chem. 2006 Aug 4;281(31):21884-21891. doi: 10.1074/jbc.M600427200. Epub 2006 Jun 9. J Biol Chem. 2006. PMID: 16766796 Free PMC article. - Mutagenic analysis in a pure molecular system shows that thioredoxin-interacting protein residue Cys247 is necessary and sufficient for a mixed disulfide formation with thioredoxin.
Fould B, Lamamy V, Guenin SP, Ouvry C, Cogé F, Boutin JA, Ferry G. Fould B, et al. Protein Sci. 2012 Sep;21(9):1323-33. doi: 10.1002/pro.2119. Epub 2012 Aug 9. Protein Sci. 2012. PMID: 22760822 Free PMC article. - Thioredoxin interacting protein: redox dependent and independent regulatory mechanisms.
Spindel ON, World C, Berk BC. Spindel ON, et al. Antioxid Redox Signal. 2012 Mar 15;16(6):587-96. doi: 10.1089/ars.2011.4137. Epub 2011 Dec 20. Antioxid Redox Signal. 2012. PMID: 21929372 Free PMC article. Review. - Nitrosative/oxidative stress conditions regulate thioredoxin-interacting protein (TXNIP) expression and thioredoxin-1 (TRX-1) nuclear localization.
Ogata FT, Batista WL, Sartori A, Gesteira TF, Masutani H, Arai RJ, Yodoi J, Stern A, Monteiro HP. Ogata FT, et al. PLoS One. 2013 Dec 20;8(12):e84588. doi: 10.1371/journal.pone.0084588. eCollection 2013. PLoS One. 2013. PMID: 24376827 Free PMC article. - TXNIP: A Double-Edged Sword in Disease and Therapeutic Outlook.
Pan M, Zhang F, Qu K, Liu C, Zhang J. Pan M, et al. Oxid Med Cell Longev. 2022 Apr 11;2022:7805115. doi: 10.1155/2022/7805115. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35450411 Free PMC article. Review.
Cited by
- The thioredoxin system: Balancing redox responses in immune cells and tumors.
Muri J, Kopf M. Muri J, et al. Eur J Immunol. 2023 Jan;53(1):e2249948. doi: 10.1002/eji.202249948. Epub 2022 Nov 17. Eur J Immunol. 2023. PMID: 36285367 Free PMC article. Review. - TXNIP-mediated crosstalk between oxidative stress and glucose metabolism.
Kim S, Ge J, Kim D, Lee JJ, Choi YJ, Chen W, Bowman JW, Foo SS, Chang LC, Liang Q, Hara D, Choi I, Kim MH, Eoh H, Jung JU. Kim S, et al. PLoS One. 2024 Feb 8;19(2):e0292655. doi: 10.1371/journal.pone.0292655. eCollection 2024. PLoS One. 2024. PMID: 38329960 Free PMC article. - Ras Suppresses TXNIP Expression by Restricting Ribosome Translocation.
Ye Z, Ayer DE. Ye Z, et al. Mol Cell Biol. 2018 Sep 28;38(20):e00178-18. doi: 10.1128/MCB.00178-18. Print 2018 Oct 15. Mol Cell Biol. 2018. PMID: 30037981 Free PMC article. - Thioredoxin-interacting protein is essential for memory T cell formation via the regulation of the redox metabolism.
Kokubo K, Hirahara K, Kiuchi M, Tsuji K, Shimada Y, Sonobe Y, Shinmi R, Hishiya T, Iwamura C, Onodera A, Nakayama T. Kokubo K, et al. Proc Natl Acad Sci U S A. 2023 Jan 10;120(2):e2218345120. doi: 10.1073/pnas.2218345120. Epub 2023 Jan 3. Proc Natl Acad Sci U S A. 2023. PMID: 36595680 Free PMC article. - Galectin-3 induces vascular smooth muscle cells calcification via AMPK/TXNIP pathway.
Tian L, Wang Y, Zhang R. Tian L, et al. Aging (Albany NY). 2022 Jun 27;14(12):5086-5096. doi: 10.18632/aging.204130. Epub 2022 Jun 27. Aging (Albany NY). 2022. PMID: 35771146 Free PMC article.
References
- Lillig C. H. & Holmgren A. Thioredoxin and related molecules--from biology to health and disease. Antioxid. Redox Signal. 9, 25–47 (2007). - PubMed
- Burke-Gaffney A., Callister M. E. & Nakamura H. Thioredoxin: friend or foe in human disease? Trends Pharmacol. Sci. 26, 398–404 (2005). - PubMed
- Grogan T. M. et al. Thioredoxin, a putative oncogene product, is overexpressed in gastric carcinoma and associated with increased proliferation and increased cell survival. Hum. Pathol. 31, 475–481 (2000). - PubMed
- Raffel J. et al. Increased expression of thioredoxin-1 in human colorectal cancer is associated with decreased patient survival. J. Lab. Clin. Med. 142, 46–51 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous