Diversity of Prdm9 zinc finger array in wild mice unravels new facets of the evolutionary turnover of this coding minisatellite - PubMed (original) (raw)
Diversity of Prdm9 zinc finger array in wild mice unravels new facets of the evolutionary turnover of this coding minisatellite
Jérôme Buard et al. PLoS One. 2014.
Abstract
In humans and mice, meiotic recombination events cluster into narrow hotspots whose genomic positions are defined by the PRDM9 protein via its DNA binding domain constituted of an array of zinc fingers (ZnFs). High polymorphism and rapid divergence of the Prdm9 gene ZnF domain appear to involve positive selection at DNA-recognition amino-acid positions, but the nature of the underlying evolutionary pressures remains a puzzle. Here we explore the variability of the Prdm9 ZnF array in wild mice, and uncovered a high allelic diversity of both ZnF copy number and identity with the caracterization of 113 alleles. We analyze features of the diversity of ZnF identity which is mostly due to non-synonymous changes at codons -1, 3 and 6 of each ZnF, corresponding to amino-acids involved in DNA binding. Using methods adapted to the minisatellite structure of the ZnF array, we infer a phylogenetic tree of these alleles. We find the sister species Mus spicilegus and M. macedonicus as well as the three house mouse (Mus musculus) subspecies to be polyphyletic. However some sublineages have expanded independently in Mus musculus musculus and M. m. domesticus, the latter further showing phylogeographic substructure. Compared to random genomic regions and non-coding minisatellites, none of these patterns appears exceptional. In silico prediction of DNA binding sites for each allele, overlap of their alignments to the genome and relative coverage of the different families of interspersed repeated elements suggest a large diversity between PRDM9 variants with a potential for highly divergent distributions of recombination events in the genome with little correlation to evolutionary distance. By compiling PRDM9 ZnF protein sequences in Primates, Muridae and Equids, we find different diversity patterns among the three amino-acids most critical for the DNA-recognition function, suggesting different diversification timescales.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
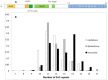
Figure 1. The C2H2 zinc finger domain of PRDM9.
(A) PRDM9 contains several identified domains: an amino-terminal region which includes a Krüppel-associated box (KRAB) domain and an SSX repression domain (SSXRD); a PR/SET domain carrying methyltransferase activity, surrounded by a zinc knuckle and a zinc finger; and a long carboxy-terminal C2H2 zinc finger array. In this example, such as observed in the mouse laboratory strain C57BL/6, the array is composed of 12 zinc fingers. (B) Size distribution of the Prdm9 ZnF arrays (number of ZnF repeats) genotyped in the three subspecies of the house mouse
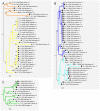
Figure 2. Inferred phylogeny of the Prdm9 ZnF domain alleles.
Taxon names on the branches include allele number, followed by the number of observations, then by taxon code (abbreviation of species name), country code, locality name and number of ZnF repeats. Numbers at the nodes indicate the level of confidence of the node (only values >0.5 reported).
Figure 3. Details of phylogenic tree.
The yellow (A), blue (B) and green (C) collapsed parts of the tree in Fig. 2 are expended and represent the different house mouse lineages. Taxon name encoding is as in Fig. 2.
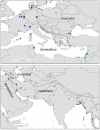
Figure 4. Geographic distribution of groups of alleles in the house mouse.
The shape of the symbols indicates subspecies (square, M. m. domesticus, circles M. m. musculus, triangles M. m. castaneus). Colors indicate lineages as in Fig. 3.
Figure 5. Simplified triplet protein variants of the Prdm9 ZnF array in wild mice.
Sequence identifiers are highlighted with colors as in the phylogenetic tree of DNA alleles in Fig. 2 and 3. Alleles of laboratory strains previously sequenced are identified as in . Each ZnF is simplified to the three most variable codons −1, 3 and 6, and separated with a dash from the next ZnF. Sequences start at the first functional C2H2 ZnF (the second repeat) and end at the last carboxy-terminal ZnF of the protein. A few remarkable stretches of zinc fingers are highlighted: some are shared between most M. musculus protein variants (QNK-QDQ, red), some are shared between the twin species spicilegus and macedonicus (QNQ-ANK-**Q-QDQ, purple), some are shared between castaneus and musculus alleles (ANQ-ESK, yellow) and some others are specific or enriched in each of domesticus (QHQ-QDK, dark blue; AVQ-AVQ, light blue), castaneus (VVQ, green), M. spretus (ADK-VNQ; QNQ-ADK, grey); M. macedonicus (QHK-QNQ, purple) and M. spicilegus (QNQ-ADK, grey) groups of alleles.
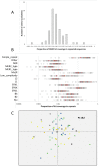
Figure 6. Predicted DNA binding sites of mouse Prdm9 ZnF alleles and dispersed repeats.
(A) Distribution among Prdm9 alleles of the proportion of the coverage of hits of the predicted recognized DNA motifs that fall in dispersed repeated sequences, as annotated on the reference mouse genome. (B) Absolute proportion of hit coverage falling in a given repeat family for each of the sequenced allele. Red cross: expected proportion if hit coverage was proportional to the coverage of the family in the genome. Red circles: median, first and third quartile of the distribution across alleles. Note the log scales. (C) Projection of the alleles on the first two axes of the Principal Component Analysis on the relative proportion of hits of each allele in the different repeated families. Symbol colors refer to lineage colors as in Fig. 2 and 3. Symbol shapes are arbitrary. PC1 absorbs 35% of the variance, and PC2 13%.
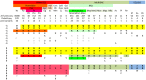
Figure 7. Patterns of diversity of amino-acids –1, 3 and 6 of PRDM9 ZnFs across taxa, species and subspecies.
Number of ZnFs units sequenced, number of Prdm9 ZnF arrays sequenced and number of protein variants are indicated for each species or subspecies of Primates, Muridae and Equids. Variant amino-acids at each of the three positions −1, 3 and 6 of the PRDM9 ZnFs are shown for each group. Variants present in every allele sequenced are in bold case and variants found in less than 10% of ZnF units are in grey case (all in normal case when one allele is available). Some variants at position −1 and position 3 are shared by most species (highlighted in yellow), others are shared by one taxon (highlighted according to the colour of the taxon). Hs: Homo sapiens; Pp: Pan paniscus (bonobo); Pt: Pan troglodytes (chimpanzee); Ptt_: P. T. troglodytes_; Ptv: P. T. verus; Pts: P. T. schweininfurthii; Gg: Gorilla gorilla; Hol: _Holobylata_e; Nl: Nomascus leucogenys (Gibbon); Cerc: Cercopithecidae; Mm: Macaca mulata (Rhesus monkey); Calli: Callitrichidae; Cj: Callithrix jacchus (Ouistiti); Gal: Galagidae; Og: Otolemur garnettii (Lemur); Mm: Mus musculus; Mmd: Mus musculus domesticus; Mmm: M. m. musculus; Mmc: M. m. castaneus; Msp: Mus spretus; Mm/s: Mus macedonicus and spicigelus; Mpy: Mus Pyromys platythrix; Mfa: Mus famulus; As: Apodemus sylvaticus; Pl: Peromyscus leucopus; Rn: Rattus norvegicus; Ef: Equus ferus; Ea: Equus asinus; Eh: Equus hippotigris. Data was gathered for Mus ZnFs from this study, for Homo sapiens ZnFs from , , , for Pan ZnFs from , , for Equids from and retrieved from GenBank for other individual alleles (Gg, Nl, Mm, Cj, Og, Apos, Perol, Rn).
Similar articles
- PRDM9, a driver of the genetic map.
Grey C, Baudat F, de Massy B. Grey C, et al. PLoS Genet. 2018 Aug 30;14(8):e1007479. doi: 10.1371/journal.pgen.1007479. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30161134 Free PMC article. Review. - Prdm9 polymorphism unveils mouse evolutionary tracks.
Kono H, Tamura M, Osada N, Suzuki H, Abe K, Moriwaki K, Ohta K, Shiroishi T. Kono H, et al. DNA Res. 2014 Jun;21(3):315-26. doi: 10.1093/dnares/dst059. Epub 2014 Jan 20. DNA Res. 2014. PMID: 24449848 Free PMC article. - The long zinc finger domain of PRDM9 forms a highly stable and long-lived complex with its DNA recognition sequence.
Striedner Y, Schwarz T, Welte T, Futschik A, Rant U, Tiemann-Boege I. Striedner Y, et al. Chromosome Res. 2017 Jun;25(2):155-172. doi: 10.1007/s10577-017-9552-1. Epub 2017 Feb 2. Chromosome Res. 2017. PMID: 28155083 Free PMC article. - Rapid Evolution of the Fine-scale Recombination Landscape in Wild House Mouse (Mus musculus) Populations.
Wooldridge LK, Dumont BL. Wooldridge LK, et al. Mol Biol Evol. 2023 Jan 4;40(1):msac267. doi: 10.1093/molbev/msac267. Mol Biol Evol. 2023. PMID: 36508360 Free PMC article. - The case of the fickle fingers: how the PRDM9 zinc finger protein specifies meiotic recombination hotspots in humans.
Ségurel L, Leffler EM, Przeworski M. Ségurel L, et al. PLoS Biol. 2011 Dec;9(12):e1001211. doi: 10.1371/journal.pbio.1001211. Epub 2011 Dec 6. PLoS Biol. 2011. PMID: 22162947 Free PMC article. Review.
Cited by
- PRDM9, a driver of the genetic map.
Grey C, Baudat F, de Massy B. Grey C, et al. PLoS Genet. 2018 Aug 30;14(8):e1007479. doi: 10.1371/journal.pgen.1007479. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30161134 Free PMC article. Review. - Meiotic recombination mirrors patterns of germline replication in mice and humans.
Pratto F, Brick K, Cheng G, Lam KG, Cloutier JM, Dahiya D, Wellard SR, Jordan PW, Camerini-Otero RD. Pratto F, et al. Cell. 2021 Aug 5;184(16):4251-4267.e20. doi: 10.1016/j.cell.2021.06.025. Epub 2021 Jul 13. Cell. 2021. PMID: 34260899 Free PMC article. - Recombination rate variation in mice from an isolated island.
Wang RJ, Gray MM, Parmenter MD, Broman KW, Payseur BA. Wang RJ, et al. Mol Ecol. 2017 Jan;26(2):457-470. doi: 10.1111/mec.13932. Epub 2016 Dec 21. Mol Ecol. 2017. PMID: 27864900 Free PMC article. - In vivo binding of PRDM9 reveals interactions with noncanonical genomic sites.
Grey C, Clément JA, Buard J, Leblanc B, Gut I, Gut M, Duret L, de Massy B. Grey C, et al. Genome Res. 2017 Apr;27(4):580-590. doi: 10.1101/gr.217240.116. Epub 2017 Mar 23. Genome Res. 2017. PMID: 28336543 Free PMC article. - The Role of KRAB-ZFPs in Transposable Element Repression and Mammalian Evolution.
Yang P, Wang Y, Macfarlan TS. Yang P, et al. Trends Genet. 2017 Nov;33(11):871-881. doi: 10.1016/j.tig.2017.08.006. Epub 2017 Sep 19. Trends Genet. 2017. PMID: 28935117 Free PMC article. Review.
References
- Handel MA, Schimenti JC (2010) Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 11: 124–136. - PubMed
- McKee BD (2009) Homolog pairing and segregation in Drosophila meiosis. Genome Dyn 5: 56–68. - PubMed
- Otto SP, Lenormand T (2002) Resolving the paradox of sex and recombination. Nat Rev Genet 3: 252–261. - PubMed
- Coop G, Przeworski M (2007) An evolutionary view of human recombination. Nat Rev Genet 8: 23–34. - PubMed
- Baudat F, de Massy B (2007) Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res 15: 565–577. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
JB and BdM are supported by the Centre National de la Recherche Scientifique, the Agence Nationale de la Recherche (09-BLAN-0269-01), and the Fondation pour la Recherche Médicale. DD is recipient of a fellowship by from the Labex « EpigenMed » program of MENRT. The project was partly funded by the Conseil Scientifique of Université Montpellier 2 (AAP2011 to PB). ER is supported by the Region Languedoc Roussillon (grant Chercheur d'Avenir), the NUMEV Labex, the MASTODONS Défi from CNRS, and by Investissements d'Avenir (grant Institut Computational Biology). The funders has no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases