Distortion of genealogical properties when the sample is very large - PubMed (original) (raw)
Distortion of genealogical properties when the sample is very large
Anand Bhaskar et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Study sample sizes in human genetics are growing rapidly, and in due course it will become routine to analyze samples with hundreds of thousands, if not millions, of individuals. In addition to posing computational challenges, such large sample sizes call for carefully reexamining the theoretical foundation underlying commonly used analytical tools. Here, we study the accuracy of the coalescent, a central model for studying the ancestry of a sample of individuals. The coalescent arises as a limit of a large class of random mating models, and it is an accurate approximation to the original model provided that the population size is sufficiently larger than the sample size. We develop a method for performing exact computation in the discrete-time Wright-Fisher (DTWF) model and compare several key genealogical quantities of interest with the coalescent predictions. For recently inferred demographic scenarios, we find that there are a significant number of multiple- and simultaneous-merger events under the DTWF model, which are absent in the coalescent by construction. Furthermore, for large sample sizes, there are noticeable differences in the expected number of rare variants between the coalescent and the DTWF model. To balance the trade-off between accuracy and computational efficiency, we propose a hybrid algorithm that uses the DTWF model for the recent past and the coalescent for the more distant past. Our results demonstrate that the hybrid method with only a handful of generations of the DTWF model leads to a frequency spectrum that is quite close to the prediction of the full DTWF model.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
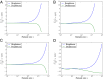
Fig. 1.
The percentage relative error in the number of singletons and doubletons between the coalescent and DTWF models, as a function of the sample size n. When the sample size is comparable to the current population size, the number of singletons predicted by the DTWF model is larger than the coalescent prediction by as much as 11%, whereas the number of doubletons predicted by the DTWF model is smaller than the coalescent prediction by about 4.8%. In model 4, we could not consider a sample size comparable to the population size (106) because of computational burden, but we expect a similar extent of deviation as in models 1–3 as n increases. Note that the _y_-axis scale for model 4 is different from that for models 1–3. (A) Model 1. (B) Model 2. (C) Model 3. (D) Model 4.
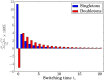
Fig. 2.
The percentage relative error, with respect to the full DTWF model, in the number of singletons and doubletons in a hybrid algorithm with switching time t s. The hybrid method uses the DTWF model for generations ≤t s and the coalescent model in generations >t s. The results are for model 3 in the case in which the sample size n is equal to the current effective population size _N_0 = 67,627. The case of t s = 0 corresponds to using the coalescent model only. This plot shows that the difference in the frequency spectrum between the full DTWF model and the hybrid algorithm decreases very rapidly as the switching time t s increases.
Similar articles
- Scaling the discrete-time Wright-Fisher model to biobank-scale datasets.
Spence JP, Zeng T, Mostafavi H, Pritchard JK. Spence JP, et al. Genetics. 2023 Nov 1;225(3):iyad168. doi: 10.1093/genetics/iyad168. Genetics. 2023. PMID: 37724741 Free PMC article. - ARGON: fast, whole-genome simulation of the discrete time Wright-fisher process.
Palamara PF. Palamara PF. Bioinformatics. 2016 Oct 1;32(19):3032-4. doi: 10.1093/bioinformatics/btw355. Epub 2016 Jun 16. Bioinformatics. 2016. PMID: 27312410 Free PMC article. - Exact coalescent for the Wright-Fisher model.
Fu YX. Fu YX. Theor Popul Biol. 2006 Jun;69(4):385-94. doi: 10.1016/j.tpb.2005.11.005. Epub 2006 Jan 19. Theor Popul Biol. 2006. PMID: 16426654 - Coalescents and genealogical structure under neutrality.
Donnelly P, Tavaré S. Donnelly P, et al. Annu Rev Genet. 1995;29:401-21. doi: 10.1146/annurev.ge.29.120195.002153. Annu Rev Genet. 1995. PMID: 8825481 Review. - [Genetic aspects of genealogy].
Tetushkin EIu. Tetushkin EIu. Genetika. 2011 Nov;47(11):1451-72. Genetika. 2011. PMID: 22332404 Review. Russian.
Cited by
- Scaling the discrete-time Wright-Fisher model to biobank-scale datasets.
Spence JP, Zeng T, Mostafavi H, Pritchard JK. Spence JP, et al. Genetics. 2023 Nov 1;225(3):iyad168. doi: 10.1093/genetics/iyad168. Genetics. 2023. PMID: 37724741 Free PMC article. - Scaling the Discrete-time Wright Fisher model to biobank-scale datasets.
Spence JP, Zeng T, Mostafavi H, Pritchard JK. Spence JP, et al. bioRxiv [Preprint]. 2023 May 22:2023.05.19.541517. doi: 10.1101/2023.05.19.541517. bioRxiv. 2023. PMID: 37293115 Free PMC article. Updated. Preprint. - Labelled histories with multifurcation and simultaneity.
Dickey EH, Rosenberg NA. Dickey EH, et al. Philos Trans R Soc Lond B Biol Sci. 2025 Feb 13;380(1919):20230307. doi: 10.1098/rstb.2023.0307. Epub 2025 Feb 20. Philos Trans R Soc Lond B Biol Sci. 2025. PMID: 39976407 - Coalescent inferences in conservation genetics: should the exception become the rule?
Montano V. Montano V. Biol Lett. 2016 Jun;12(6):20160211. doi: 10.1098/rsbl.2016.0211. Biol Lett. 2016. PMID: 27330172 Free PMC article. Review. - High burden of private mutations due to explosive human population growth and purifying selection.
Gao F, Keinan A. Gao F, et al. BMC Genomics. 2014;15 Suppl 4(Suppl 4):S3. doi: 10.1186/1471-2164-15-S4-S3. Epub 2014 May 20. BMC Genomics. 2014. PMID: 25056720 Free PMC article.
References
- Takahata N. Allelic genealogy and human evolution. Mol Biol Evol. 1993;10(1):2–22. - PubMed
- Erlich HA, Bergström TF, Stoneking M, Gyllensten U. HLA sequence polymorphism and the origin of humans. Science. 1996;274(5292):1552–1554. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 GM108805/GM/NIGMS NIH HHS/United States
- R01 HG003229/HG/NHGRI NIH HHS/United States
- R01-HG003229/HG/NHGRI NIH HHS/United States
- R01 GM094402/GM/NIGMS NIH HHS/United States
- R01-GM094402/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous