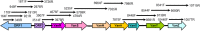Outbreak of vancomycin-susceptible Enterococcus faecium containing the wild-type vanA gene - PubMed (original) (raw)
Outbreak of vancomycin-susceptible Enterococcus faecium containing the wild-type vanA gene
Tom A Szakacs et al. J Clin Microbiol. 2014 May.
Abstract
Accurate detection of vancomycin-resistant enterococci (VRE) is essential in preventing transmission in health care settings. Chromogenic media are widely used for screening VRE because of fast turnaround times (TAT) and high sensitivity. We report an outbreak of Enterococcus faecium bearing vanA yet susceptible to vancomycin (vancomycin-variable Enterococcus [VVE]). Between October 2009 to March 2011, clinical and screening specimens (n=14,747) were screened for VRE using VRE-selective medium and/or PCR. VVE isolates were genotyped to determine relatedness. Plasmids from these isolates were characterized by sequencing. Overall, 52 VVE isolates were identified, comprising 15% of all VRE isolates identified. Isolates demonstrated growth on Brilliance VRE agar (Oxoid) at 24 h of incubation but did not grow on brain heart infusion agar with 6 μg/ml vancomycin (Oxoid) or bile esculin azide agar with 6 μg/ml vancomycin (Oxoid) and were susceptible to vancomycin. Genotyping of 20 randomly selected VVE isolates revealed that 15/20 were identical, while 5 were highly related. PCR of the VVE transposon confirmed the presence of vanHAXY gene cluster; however, vanS (sensor) and vanR (regulator) genes were absent. The outbreak was controlled through routine infection control measures. We report an emergence of a fit strain of E. faecium containing vanA yet susceptible to vancomycin. Whether this new strain represents VRE has yet to be determined; however, unique testing procedures are required for reliable identification of VVE.
Figures
FIG 1
Genetic map and PCR amplification position of Tn_1546_. A schematic diagram of overlapping primers used to amplify and detect the transposon is shown.
FIG 2
Genetic map of the full-length 31,136-bp plasmid, pF856. Coding regions are represented by arrows indicating the direction of transcription. Insertion sequence elements and type 1 RM-S are shown in gray, and Tn_1546_ transposon elements are in white. Putative ORFs are labeled according to the most significant homologue (pS177) in the public databases.
FIG 3
Pulse-field gel electrophoresis (PFGE) patterns of VVE strains. Chromosomal DNA was digested with SmaI.
Similar articles
- Surveillance of vancomycin-resistant enterococci reveals shift in dominating clusters from vanA to vanB Enterococcus faecium clusters, Denmark, 2015 to 2022.
Hammerum AM, Karstensen KT, Roer L, Kaya H, Lindegaard M, Porsbo LJ, Kjerulf A, Pinholt M, Holzknecht BJ, Worning P, Nielsen KL, Hansen SGK, Clausen M, Søndergaard TS, Dzajic E, Østergaard C, Wang M, Koch K, Hasman H. Hammerum AM, et al. Euro Surveill. 2024 Jun;29(23):2300633. doi: 10.2807/1560-7917.ES.2024.29.23.2300633. Euro Surveill. 2024. PMID: 38847117 Free PMC article. - Prevalence of vancomycin-variable Enterococcus faecium (VVE) among vanA-positive sterile site isolates and patient factors associated with VVE bacteremia.
Kohler P, Eshaghi A, Kim HC, Plevneshi A, Green K, Willey BM, McGeer A, Patel SN; Toronto Invasive Bacterial Diseases Network (TIBDN). Kohler P, et al. PLoS One. 2018 Mar 22;13(3):e0193926. doi: 10.1371/journal.pone.0193926. eCollection 2018. PLoS One. 2018. PMID: 29566004 Free PMC article. - Prevalence and Molecular Characterization of Vancomycin Variable Enterococcus faecium Isolated From Clinical Specimens.
Yoo IY, Kwon JA, Lee M, Jung SH, Kim JO, Ha SI, Park YJ. Yoo IY, et al. Ann Lab Med. 2024 Sep 1;44(5):450-454. doi: 10.3343/alm.2023.0430. Epub 2024 Mar 13. Ann Lab Med. 2024. PMID: 38475872 Free PMC article. - Thirty years of VRE in Germany - "expect the unexpected": The view from the National Reference Centre for Staphylococci and Enterococci.
Werner G, Neumann B, Weber RE, Kresken M, Wendt C, Bender JK; VRE study group. Werner G, et al. Drug Resist Updat. 2020 Dec;53:100732. doi: 10.1016/j.drup.2020.100732. Epub 2020 Oct 27. Drug Resist Updat. 2020. PMID: 33189998 Review. - Vancomycin-resistant Enterococcus faecium and the emergence of new sequence types associated with hospital infection.
O'Toole RF, Leong KWC, Cumming V, Van Hal SJ. O'Toole RF, et al. Res Microbiol. 2023 May;174(4):104046. doi: 10.1016/j.resmic.2023.104046. Epub 2023 Feb 27. Res Microbiol. 2023. PMID: 36858192 Review.
Cited by
- Occurrence of vanHAX and Related Genes beyond the Actinobacteria Phylum.
Yushchuk O, Binda E, Fedorenko V, Marinelli F. Yushchuk O, et al. Genes (Basel). 2022 Oct 27;13(11):1960. doi: 10.3390/genes13111960. Genes (Basel). 2022. PMID: 36360197 Free PMC article. - Investigation of vancomycin resistant Enterococcus faecium outbreak in neonatal intensive care unit.
Cilo BD, Ağca H, Efe K, Sınırtaş M, Çelebi S, Özkan H, Köksal N, Hacımustafaoğlu M, Özakın C. Cilo BD, et al. Int J Clin Exp Med. 2014 Dec 15;7(12):5342-7. eCollection 2014. Int J Clin Exp Med. 2014. PMID: 25664041 Free PMC article. - Surveillance of vancomycin-resistant enterococci reveals shift in dominating clones and national spread of a vancomycin-variable vanA Enterococcus faecium ST1421-CT1134 clone, Denmark, 2015 to March 2019.
Hammerum AM, Justesen US, Pinholt M, Roer L, Kaya H, Worning P, Nygaard S, Kemp M, Clausen ME, Nielsen KL, Samulioniené J, Kjærsgaard M, Østergaard C, Coia J, Søndergaard TS, Gaini S, Schønning K, Westh H, Hasman H, Holzknecht BJ. Hammerum AM, et al. Euro Surveill. 2019 Aug;24(34):1900503. doi: 10.2807/1560-7917.ES.2019.24.34.1900503. Euro Surveill. 2019. PMID: 31456560 Free PMC article. - A Functional Metagenomic Analysis of Tetracycline Resistance in Cheese Bacteria.
Flórez AB, Vázquez L, Mayo B. Flórez AB, et al. Front Microbiol. 2017 May 24;8:907. doi: 10.3389/fmicb.2017.00907. eCollection 2017. Front Microbiol. 2017. PMID: 28596758 Free PMC article. - Comparative genomics of Clostridium bolteae and Clostridium clostridioforme reveals species-specific genomic properties and numerous putative antibiotic resistance determinants.
Dehoux P, Marvaud JC, Abouelleil A, Earl AM, Lambert T, Dauga C. Dehoux P, et al. BMC Genomics. 2016 Oct 21;17(1):819. doi: 10.1186/s12864-016-3152-x. BMC Genomics. 2016. PMID: 27769168 Free PMC article.
References
- Ofner-Agostini M, Johnston BL, Simor AE, Embil J, Matlow A, Mulvey M, Ormiston D, Conly J, Canadian Nosocomial Infection Surveillance Program 2008. Vancomycin-resistant enterococci in Canada: results from the Canadian nosocomial infection surveillance program, 1999–2005. Infect. Control Hosp. Epidemiol. 29:271–274. 10.1086/528812 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources