Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels - PubMed (original) (raw)
. 2014 Jul;13(7):1690-704.
doi: 10.1074/mcp.M113.036392. Epub 2014 Apr 9.
Feng Yang 2, Tao Liu 2, D R Mani 3, Vladislav A Petyuk 2, Michael A Gillette 3, Karl R Clauser 3, Jana W Qiao 3, Marina A Gritsenko 2, Ronald J Moore 2, Douglas A Levine 4, Reid Townsend 5, Petra Erdmann-Gilmore 5, Jacqueline E Snider 5, Sherri R Davies 5, Kelly V Ruggles 6, David Fenyo 6, R Thomas Kitchens 5, Shunqiang Li 5, Narciso Olvera 4, Fanny Dao 4, Henry Rodriguez 7, Daniel W Chan 8, Daniel Liebler 9, Forest White 10, Karin D Rodland 2, Gordon B Mills 11, Richard D Smith 2, Amanda G Paulovich 12, Matthew Ellis 5, Steven A Carr 1
Affiliations
- PMID: 24719451
- PMCID: PMC4083109
- DOI: 10.1074/mcp.M113.036392
Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels
Philipp Mertins et al. Mol Cell Proteomics. 2014 Jul.
Abstract
Protein abundance and phosphorylation convey important information about pathway activity and molecular pathophysiology in diseases including cancer, providing biological insight, informing drug and diagnostic development, and guiding therapeutic intervention. Analyzed tissues are usually collected without tight regulation or documentation of ischemic time. To evaluate the impact of ischemia, we collected human ovarian tumor and breast cancer xenograft tissue without vascular interruption and performed quantitative proteomics and phosphoproteomics after defined ischemic intervals. Although the global expressed proteome and most of the >25,000 quantified phosphosites were unchanged after 60 min, rapid phosphorylation changes were observed in up to 24% of the phosphoproteome, representing activation of critical cancer pathways related to stress response, transcriptional regulation, and cell death. Both pan-tumor and tissue-specific changes were observed. The demonstrated impact of pre-analytical tissue ischemia on tumor biology mandates caution in interpreting stress-pathway activation in such samples and motivates reexamination of collection protocols for phosphoprotein analysis.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
Fig. 1.
Quantitative proteome and phosphoproteome analysis of human ovarian tumors and xenograft breast tumors subjected to controlled ischemia. A, experimental design to study effects of post-excision delay time before freezing across four time points. After excision, tumor samples were cut into four equal pieces and incubated for the indicated times at room temperature before freezing. A total of four different ovarian tumors and three pooled breast cancer xenograft samples for the basal and for the luminal subtype were analyzed. All of these samples were biologically distinct and can be considered as biological replicates. B, quantitative proteomics and phosphoproteomics workflow using 4-plex iTRAQ labeling. Tumor samples were cryofractured and proteins were extracted with urea lysis buffer prior to digestion into peptides using trypsin. Peptide samples derived at four different ischemic time points were labeled using iTRAQ reagents, mixed equably, and separated using high-pH reversed-phase chromatography. Fractions were combined in a noncontiguous way into 24 fractions for proteome analysis (5% of the total material) and 12 fractions for phosphoproteome analysis (95% of the total material). Ovarian cancer samples were analyzed on an LTQ-Orbitrap Velos, and xenograft breast cancer samples were analyzed on a Q Exactive mass spectrometer. Phosphosite and protein identification and quantification were achieved using Spectrum Mill.
Fig. 2.
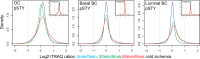
Global proteome analysis revealed no changes in protein abundance level, but specific alterations in the phosphoproteome induced by ischemia were observed. Density plots are shown for averaged phosphosite iTRAQ ratios (labeled with “pSTY”) and protein iTRAQ ratios (small insets) for the ovarian cancer (OC) samples and the basal-like and luminal breast cancer (BC) samples. Only phosphosites/proteins were plotted that were quantified in at least three ovarian tumors and at least two basal-like or luminal tumor samples.
Fig. 3.
Temporal dynamics of phosphorylation changes resulting from cold ischemia. A, fuzzy c-means clustering of temporal profiles for all regulated phosphosites observed in the ovarian and breast cancer samples. We detected six up-regulated (U) clusters, which were further grouped pairwise into early (U1), middle (U2), and late clusters (U3), and three down-regulated (D1, D2, D3) clusters using a fuzzyfication parameter m = 1.6. Phosphosites were assigned to each cluster with a membership value α > 0.7. T1/2 indicates the median over all half-maximum time points for all phosphosites within a cluster. Half-maximum time points were determined via first-order kinetic modeling analysis. B, number of regulated phosphosites assigned to each cluster for the ovarian and breast cancer tumors. C, gene enrichment analysis of regulated human phosphoproteins across early, middle, and late clusters. We used DAVID Bioinformatics Resources 6.7 (39) to test for enrichment of GO BPs in each cluster relative to a list of all proteins containing nonregulated sites using a modified Fisher's exact test (EASE score). GO BP categories with p < 0.01 and a minimum occurrence of ≥10 genes/proteins were called significant. p values were −Log10 transformed, and the transformed values for each annotation were plotted as a heat map in Gene-E.
Fig. 4.
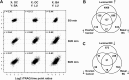
Ischemia induced common phosphorylation events in ovarian and basal-like/luminal breast cancer tumor samples. A, scatter plots of averaged phosphosite ratios (5:0, 30:0, and 60:0) over at least three ovarian tumors (OC) and at least two basal-like (BA) or luminal (LU) breast cancer samples. X and Y indicate which tumor type is plotted on the respective axis. Intertumor Pearson correlation coefficients are indicated for each tumor type comparison. B, C, Venn diagrams of the overlap of up-regulated (B) and down-regulated (C) phosphosites between ovarian and basal-like/luminal breast cancer samples.
Fig. 5.
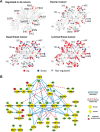
Stress-response kinases were affected by cold ischemia. A, kinases detected with and without regulated phosphosites were mapped in a dendrogram of the human kinome (40) using KinomeCluster. Ischemia-regulated kinases were observed across all major kinase subfamilies. B, protein–protein interaction and kinase-substrate network of phosphoproteins commonly regulated in all analyzed ovarian and breast cancer samples. High confidence, experimentally validated protein–protein interaction information was obtained from the STRING protein interaction database (26). Kinase–substrate relationships were derived from the PhosphoSite database (27). Protein–protein interaction and site-specific kinase–phosphosite relationships were illustrated using Cytoscape (41), with blue edges indicating protein–protein interactions and red dashed arrows indicating kinase–substrate relationships. Up-regulated phosphosites are shown in green, and down-regulated sites are in red. Kinases are depicted as squares, whereas all other proteins appear as circles.
Similar articles
- Delayed times to tissue fixation result in unpredictable global phosphoproteome changes.
Gündisch S, Grundner-Culemann K, Wolff C, Schott C, Reischauer B, Machatti M, Groelz D, Schaab C, Tebbe A, Becker KF. Gündisch S, et al. J Proteome Res. 2013 Oct 4;12(10):4424-34. doi: 10.1021/pr400451z. Epub 2013 Sep 17. J Proteome Res. 2013. PMID: 23984901 - Quantitative proteomic and phosphoproteomic profiling of ischemic myocardial stunning in swine.
Wang X, Shen X, Weil BR, Young RF, Canty JM, Qu J. Wang X, et al. Am J Physiol Heart Circ Physiol. 2020 May 1;318(5):H1256-H1271. doi: 10.1152/ajpheart.00713.2019. Epub 2020 Mar 30. Am J Physiol Heart Circ Physiol. 2020. PMID: 32223553 Free PMC article. - Phosphoproteomics reveals network rewiring to a pro-adhesion state in annexin-1-deficient mammary epithelial cells.
Alli-Shaik A, Wee S, Lim LHK, Gunaratne J. Alli-Shaik A, et al. Breast Cancer Res. 2017 Dec 12;19(1):132. doi: 10.1186/s13058-017-0924-4. Breast Cancer Res. 2017. PMID: 29233185 Free PMC article. - Illuminating the dark phosphoproteome.
Needham EJ, Parker BL, Burykin T, James DE, Humphrey SJ. Needham EJ, et al. Sci Signal. 2019 Jan 22;12(565):eaau8645. doi: 10.1126/scisignal.aau8645. Sci Signal. 2019. PMID: 30670635 Review. - Phosphoproteomics.
Ozlu N, Akten B, Timm W, Haseley N, Steen H, Steen JAJ. Ozlu N, et al. Wiley Interdiscip Rev Syst Biol Med. 2010 May-Jun;2(3):255-276. doi: 10.1002/wsbm.41. Wiley Interdiscip Rev Syst Biol Med. 2010. PMID: 20836028 Review.
Cited by
- A proteomics approach to study mouse long bones: examining baseline differences and mechanical loading-induced bone formation in young-adult and old mice.
Chermside-Scabbo CJ, Shuster JT, Erdmann-Gilmore P, Tycksen E, Zhang Q, Townsend RR, Silva MJ. Chermside-Scabbo CJ, et al. Aging (Albany NY). 2024 Oct 12;16(19):12726-12768. doi: 10.18632/aging.206131. Epub 2024 Oct 12. Aging (Albany NY). 2024. PMID: 39400554 Free PMC article. - PCPE2: Expression of multifunctional extracellular glycoprotein associated with diverse cellular functions.
Thomas MJ, Xu H, Wang A, Beg MA, Sorci-Thomas MG. Thomas MJ, et al. J Lipid Res. 2024 Oct 5;65(11):100664. doi: 10.1016/j.jlr.2024.100664. Online ahead of print. J Lipid Res. 2024. PMID: 39374805 Free PMC article. Review. - Tumor specimen cold ischemia time impacts molecular cancer drug target discovery.
von der Heyde S, Raman N, Gabelia N, Matias-Guiu X, Yoshino T, Tsukada Y, Melino G, Marshall JL, Wellstein A, Juhl H, Landgrebe J. von der Heyde S, et al. Cell Death Dis. 2024 Sep 26;15(9):691. doi: 10.1038/s41419-024-07090-x. Cell Death Dis. 2024. PMID: 39327466 Free PMC article. - CoDIAC: A comprehensive approach for interaction analysis reveals novel insights into SH2 domain function and regulation.
Kandoor A, Martinez G, Hitchcock JM, Angel S, Campbell L, Rizvi S, Naegle KM. Kandoor A, et al. bioRxiv [Preprint]. 2024 Jul 22:2024.07.18.604100. doi: 10.1101/2024.07.18.604100. bioRxiv. 2024. PMID: 39091881 Free PMC article. Preprint. - Pseudophosphorylation of single residues of the J-domain of DNAJA2 regulates the holding/folding balance of the Hsc70 system.
Velasco-Carneros L, Bernardo-Seisdedos G, Maréchal JD, Millet O, Moro F, Muga A. Velasco-Carneros L, et al. Protein Sci. 2024 Aug;33(8):e5105. doi: 10.1002/pro.5105. Protein Sci. 2024. PMID: 39012012 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- U24 CA159988/CA/NCI NIH HHS/United States
- P50 CA068438/CA/NCI NIH HHS/United States
- U54CA112970/CA/NCI NIH HHS/United States
- U24CA159988/CA/NCI NIH HHS/United States
- UL1 RR024992/RR/NCRR NIH HHS/United States
- U24CA160036/CA/NCI NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- P50 CA083639/CA/NCI NIH HHS/United States
- U24CA160035/CA/NCI NIH HHS/United States
- P30CA091842/CA/NCI NIH HHS/United States
- U24 CA160034/CA/NCI NIH HHS/United States
- P30 CA014051/CA/NCI NIH HHS/United States
- CA016672/CA/NCI NIH HHS/United States
- P01CA099031/CA/NCI NIH HHS/United States
- U54 CA112970/CA/NCI NIH HHS/United States
- P30 CA16672/CA/NCI NIH HHS/United States
- U24 CA160036/CA/NCI NIH HHS/United States
- U24 CA160019/CA/NCI NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- U24 CA160035/CA/NCI NIH HHS/United States
- P01 CA099031/CA/NCI NIH HHS/United States
- U24CA160034/CA/NCI NIH HHS/United States
- 3P50 CA68438/CA/NCI NIH HHS/United States
- U24CA160019/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous