Hantavirus Gn and Gc envelope glycoproteins: key structural units for virus cell entry and virus assembly - PubMed (original) (raw)
Review
Hantavirus Gn and Gc envelope glycoproteins: key structural units for virus cell entry and virus assembly
Nicolás Cifuentes-Muñoz et al. Viruses. 2014.
Abstract
In recent years, ultrastructural studies of viral surface spikes from three different genera within the Bunyaviridae family have revealed a remarkable diversity in their spike organization. Despite this structural heterogeneity, in every case the spikes seem to be composed of heterodimers formed by Gn and Gc envelope glycoproteins. In this review, current knowledge of the Gn and Gc structures and their functions in virus cell entry and exit is summarized. During virus cell entry, the role of Gn and Gc in receptor binding has not yet been determined. Nevertheless, biochemical studies suggest that the subsequent virus-membrane fusion activity is accomplished by Gc. Further, a class II fusion protein conformation has been predicted for Gc of hantaviruses, and novel crystallographic data confirmed such a fold for the Rift Valley fever virus (RVFV) Gc protein. During virus cell exit, the assembly of different viral components seems to be established by interaction of Gn and Gc cytoplasmic tails (CT) with internal viral ribonucleocapsids. Moreover, recent findings show that hantavirus glycoproteins accomplish important roles during virus budding since they self-assemble into virus-like particles. Collectively, these novel insights provide essential information for gaining a more detailed understanding of Gn and Gc functions in the early and late steps of the hantavirus infection cycle.
Figures
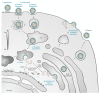
Figure 1
Schematic Representation of Hantavirus Particles. (A) Representation of TULV and HTNV particles. Surface spike organizations are based on the cryo-ET maps of each virus. (B) Amplified view of the spike arrangements of HTNV and TULV represented in (A). Spike projections were drawn based on a vertical slice through the volume of the electron density of cryo-ET maps obtained by Huiskonen and colleagues [17] and Battisti and colleagues [16], respectively. The maps were downloaded from EMDataBank, accession numbers 1704 (TULV) and 2056 (HTNV) and surface projections rendered in the Chimera program [18].
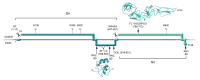
Figure 2
Functions of Hantavirus Envelope Glycoproteins during Virus Life Cycle. Once hantavirus particles attach to cell surface receptors by one, or both glycoproteins (1) they are uptaken by clathrin-mediated endocytosis (2a) or alternatively by other pathways that do not involve clathrin (2b). After virus particle uptake, the viral glycoproteins dissociate at one point from cellular receptors (3). Virus particles traffic down through the endocytic pathway until the low pH of endosomes, and possibly other cellular factors, trigger the Gc-mediated virus-cell membrane fusion process, releasing RNCs to the cytosol (4). After the transcription/replication of the viral genome and the synthesis of new viral proteins have taken place (5), the glycoproteins accumulate at internal membranes, where they can mediate virus assembly and budding (6a) and egress through the secretory pathway (7). Alternatively, new viruses may assemble and bud directly from the plasma membrane (6b). E.E. indicates early endosome, L.E. indicates late endosome.
Figure 3
Schematic Representation of Hantavirus Glycoprotein Processing and Functions. The Gn (dark cyan) and Gc (light cyan) glycoprotein ectodomains, transmembrane regions and endodomains are represented according to their location relative to the membrane (horizontal grey line). The Signal peptide (SP) and WAASA sequences indicate the cleavage sites within GPC. N represents the location of asparagine residues that are likely to carry glycosylations; the numbers following these letters indicate the corresponding residue number within GPC of ANDV. ZF 1 + 2 indicates the location of the two zinc finger domains, RNC-BS indicates the suggested ribonucleocapsid binding sites. The motif YxxL represents residues involved in ubiquitination. FL indicates the location of the putative fusion loop of ANDV Gc. The molecular structure of the ANDV Gn-CT zinc finger domains (dark cyan) was obtained from the NCBI databank, PDBid: 2K9H [24]. The molecular model for the ANDV Gc fusion protein ectodomain was developed previously [29]. All ribbon diagramsof molecular structures were rendered in the PyMOL Molecular Graphics System [61].
Similar articles
- Molecular organization and dynamics of the fusion protein Gc at the hantavirus surface.
Bignon EA, Albornoz A, Guardado-Calvo P, Rey FA, Tischler ND. Bignon EA, et al. Elife. 2019 Jun 10;8:e46028. doi: 10.7554/eLife.46028. Elife. 2019. PMID: 31180319 Free PMC article. - The Role of Phlebovirus Glycoproteins in Viral Entry, Assembly and Release.
Spiegel M, Plegge T, Pöhlmann S. Spiegel M, et al. Viruses. 2016 Jul 21;8(7):202. doi: 10.3390/v8070202. Viruses. 2016. PMID: 27455305 Free PMC article. Review. - Structural Transitions of the Conserved and Metastable Hantaviral Glycoprotein Envelope.
Rissanen I, Stass R, Zeltina A, Li S, Hepojoki J, Harlos K, Gilbert RJC, Huiskonen JT, Bowden TA. Rissanen I, et al. J Virol. 2017 Oct 13;91(21):e00378-17. doi: 10.1128/JVI.00378-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28835498 Free PMC article. - Two Point Mutations in Old World Hantavirus Glycoproteins Afford the Generation of Highly Infectious Recombinant Vesicular Stomatitis Virus Vectors.
Slough MM, Chandran K, Jangra RK. Slough MM, et al. mBio. 2019 Jan 8;10(1):e02372-18. doi: 10.1128/mBio.02372-18. mBio. 2019. PMID: 30622188 Free PMC article. - Hantavirus structure--molecular interactions behind the scene.
Hepojoki J, Strandin T, Lankinen H, Vaheri A. Hepojoki J, et al. J Gen Virol. 2012 Aug;93(Pt 8):1631-1644. doi: 10.1099/vir.0.042218-0. Epub 2012 May 23. J Gen Virol. 2012. PMID: 22622328 Review.
Cited by
- The viral nucleocapsid protein and the human RNA-binding protein Mex3A promote translation of the Andes orthohantavirus small mRNA.
Vera-Otarola J, Castillo-Vargas E, Angulo J, Barriga FM, Batlle E, Lopez-Lastra M. Vera-Otarola J, et al. PLoS Pathog. 2021 Sep 21;17(9):e1009931. doi: 10.1371/journal.ppat.1009931. eCollection 2021 Sep. PLoS Pathog. 2021. PMID: 34547046 Free PMC article. - Complete Genome Sequence of a Novel Mutation of Seoul Virus Isolated from Suncus murinus in the Fujian Province of China.
Wang YP, Zhang XL, Zhang JM, Cao XM, Han H, Huang YN, Wang HL, Chen M, Gao B, Yao LS. Wang YP, et al. Genome Announc. 2015 Apr 2;3(2):e00075-15. doi: 10.1128/genomeA.00075-15. Genome Announc. 2015. PMID: 25838472 Free PMC article. - Haploid Genetic Screen Reveals a Profound and Direct Dependence on Cholesterol for Hantavirus Membrane Fusion.
Kleinfelter LM, Jangra RK, Jae LT, Herbert AS, Mittler E, Stiles KM, Wirchnianski AS, Kielian M, Brummelkamp TR, Dye JM, Chandran K. Kleinfelter LM, et al. mBio. 2015 Jun 30;6(4):e00801. doi: 10.1128/mBio.00801-15. mBio. 2015. PMID: 26126854 Free PMC article. - Chikungunya E2 Protein Produced in E. coli and HEK293-T Cells-Comparison of Their Performances in ELISA.
Bagno FF, Godói LC, Figueiredo MM, Sérgio SAR, Moraes TFS, Salazar NC, Kim YC, Reyes-Sandoval A, da Fonseca FG. Bagno FF, et al. Viruses. 2020 Aug 26;12(9):939. doi: 10.3390/v12090939. Viruses. 2020. PMID: 32858804 Free PMC article. - Self-association and subcellular localization of Puumala hantavirus envelope proteins.
Sperber HS, Welke RW, Petazzi RA, Bergmann R, Schade M, Shai Y, Chiantia S, Herrmann A, Schwarzer R. Sperber HS, et al. Sci Rep. 2019 Jan 24;9(1):707. doi: 10.1038/s41598-018-36879-y. Sci Rep. 2019. PMID: 30679542 Free PMC article.
References
- Elliott R.M. Molecular biology of the Bunyaviridae. J. Gen. Virol. 1990;71:501–522. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous