A high-coverage shRNA screen identifies TMEM129 as an E3 ligase involved in ER-associated protein degradation - PubMed (original) (raw)
Michael C Bassik 2, Rutger D Luteijn 1, Cornelia M Voorburg 1, Mirjam A M Lohuis 1, Elisabeth Kremmer 3, Rob C Hoeben 4, Emily M LeProust 5, Siyuan Chen 5, Hanneke Hoelen 1, Maaike E Ressing 6, Weronika Patena 7, Jonathan S Weissman 8, Michael T McManus 9, Emmanuel J H J Wiertz 10, Robert Jan Lebbink 10
Affiliations
- PMID: 24807418
- PMCID: PMC4024746
- DOI: 10.1038/ncomms4832
Free PMC article
A high-coverage shRNA screen identifies TMEM129 as an E3 ligase involved in ER-associated protein degradation
Michael L van de Weijer et al. Nat Commun. 2014.
Free PMC article
Abstract
Misfolded ER proteins are retrotranslocated into the cytosol for degradation via the ubiquitin-proteasome system. The human cytomegalovirus protein US11 exploits this ER-associated protein degradation (ERAD) pathway to downregulate HLA class I molecules in virus-infected cells, thereby evading elimination by cytotoxic T-lymphocytes. US11-mediated degradation of HLA class I has been instrumental in the identification of key components of mammalian ERAD, including Derlin-1, p97, VIMP and SEL1L. Despite this, the process governing retrotranslocation of the substrate is still poorly understood. Here using a high-coverage genome-wide shRNA library, we identify the uncharacterized protein TMEM129 and the ubiquitin-conjugating E2 enzyme UBE2J2 to be essential for US11-mediated HLA class I downregulation. TMEM129 is an unconventional C4C4-type RING finger E3 ubiquitin ligase that resides within a complex containing various other ERAD components, including Derlin-1, Derlin-2, VIMP and p97, indicating that TMEM129 is an integral part of the ER-resident dislocation complex mediating US11-induced HLA class I degradation.
Conflict of interest statement
E.M.L. and S.C. are employed by Agilent Technologies, Inc., and Agilent reagents are used in the research presented in this article. All other authors declare no competing financial interests.
Figures
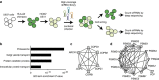
Figure 1. Pooled high-coverage RNAi screen for players involved in US11-mediated ERAD of HLA class I molecules.
(a) Experimental strategy: U937 cells were transduced with an eGFP-Myc-HLA-A2 chimaera and subsequently transduced with an HCMV US11-expression vector. The resulting cells were cloned and displayed low total eGFP-Myc-HLA-A2 expression as assessed by eGFP expression, and low cell-surface expression of the chimaeric (as assessed by anti-Myc cell-surface stain) and endogenous HLA class I alleles. The cells were infected with the pooled genome-wide high-coverage shRNA library and subjected to flow cytometry sorting at 6 dpi to select for eGFPbright cells and eGFPdim control cells. The frequency of shRNA-encoding constructs in each subpopulation was determined by deep sequencing. (b) GO-term enrichment analysis for hits from the screen was assessed using the Database for Annotation, Visualization and Integrated Discovery (DAVID). Based on the frequency in the treated and untreated subpopulations, a hit list was established covering genes that were shared among the top 100 enriched genes from two independent screens (Table 1). (c,d) Network analysis on the selected genes identified two clusters consisting of genes present in the COPI complex (c) and proteasome-associated genes (d).
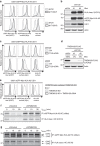
Figure 2. TMEM129 is crucial for US11-mediated HLA class I downregulation.
(a) Depletion of endogenous TMEM129 by shRNAs induces potent rescue of eGFP-Myc-HLA-A2 and endogenous HLA-A3 in U937 eGFP-Myc-HLA-A2 US11 cells. ShRNAs targeting TMEM129 (black histograms) or control shRNAs (grey histograms) were introduced. Surface endogenous HLA-A3 and surface (Myc) and total (eGFP) eGFP-Myc-HLA-A2 were assessed by flow cytometry (7 dpi). An assessment of TMEM129 levels upon shRNA depletion is presented in Fig. 2d. (b) Immunoblot analysis of endogenous HLA class I, eGFP-Myc-HLA-A2, US11, and loading control Actin in mock- (shCtrl) and TMEM129-depleted cells (shTMEM129 no. 1-2). (c) Rescue of HLA class I upon TMEM129 depletion is reversed upon overexpression of TMEM129. A TMEM129-expressing vector (black-lined histograms) or an empty vector (grey-lined histograms) were stably introduced in TMEM129- (shT129, upper panels) or mock-depleted (shCtrl, lower panels) cells, after which surface endogenous HLA-A3, and surface (Myc) and total (eGFP) eGFP-Myc-HLA-A2 were assessed by flow cytometry. (d) Immunoblot analysis of shRNA-mediated downregulation of overexpressed TMEM129-FLAG. Percentages indicate expression levels compared with mock (shCtrl) depletion normalized against Actin levels. Similar results were obtained for additional TMEM129-targeting shRNAs (Supplementary Fig. 1b–d). Depletion of endogenous TMEM129 by the first shRNA is depicted in Supplementary Fig. 9c). (e) CRISPR/Cas-mediated knockout of endogenous TMEM129 induces potent rescue of HLA class I. Surface endogenous HLA-A3, and surface (Myc) plus total (eGFP) expression of eGFP-Myc-HLA-A2 were assessed by flow cytometry in control cells (dashed histogram), TMEM129-null cells (grey-lined histograms) and TMEM129-null cells with reconstituted TMEM129 expression (black histograms). A clonal TMEM129-null cell line was established and stained; additional clones are depicted in Supplementary Fig. 2a,b. (f) Mock- (shControl) and TMEM129-depleted U937 eGFP-Myc-HLA-A2 US11 cells were subjected to a pulse chase analysis. Cells were radioactively labelled for 10 min, and chased for the indicated timeframes. Subsequently, HLA class I HCs were immunoprecipitated from lysates using indicated antibodies. (g) TMEM129 depletion in the presence of a proteasome inhibitor abrogates dislocation of HLA class I. Similar experiment as in f, albeit in the presence of the proteasome inhibitor MG132 (20 μM) immediately before and during the experiment. N-linked glycosylated (+CHO) and deglycosylated (-CHO) HLA class I HCs are indicated. The asterisk indicates a non-specific background band.
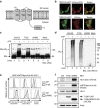
Figure 3. TMEM129 is an E3 ubiquitin ligase essential for US11-mediated HLA class I downregulation.
(a) Predicted topology of TMEM129 using the TOPCONS prediction server. TM: transmembrane domain; RING: RING domain. The N- and C terminus of the protein are depicted. (b) TMEM129 localizes to the ER. MelJuSo cells stably co-expressing TMEM129-eGFP with either mCherry (cytosolic and nuclear) or mCherry-KDEL (ER-localized) were subjected to fluorescent confocal microscopy to assess colocalization of TMEM129-eGFP with either marker. White bars represent 10 μm. (c) TMEM129 is an E3 ubiquitin ligase. In vitro ubiquitination assays were performed by using purified ubiquitin, E1 enzyme UBA1, E2 enzyme UBE2D3 and E3 enzyme MuRF1 (ctrl E3), immunoprecipitated TMEM129-FLAG-ST2 (T129) or TMEM129ΔRING-FLAG-ST2 (ΔRING), in the absence or presence of ATP. Purified S5a was used as a substrate. Immunoblot analysis was performed using an anti-S5a antibody to visualize poly-ubiquitinated S5a. Addition of the tags did not interfere with TMEM129 function (see Supplementary Fig. 10c). (d) The experiment was performed as in Fig. 3c, although a specific substrate was omitted from the reaction. Immunoblot analysis was performed using the anti-ubiquitin P4D1 mAb to visualize polyubiquitin (Ub_n_). (e) The TMEM129 RING domain is essential for US11-mediated HLA class I downregulation. Flow cytometry analysis of endogenous surface HLA-A3, and surface (Myc) and total (eGFP) eGFP-Myc-HLA-A2 in U937 eGFP-Myc-HLA-A2 cells expressing US11 and co-expressing either TMEM129 (T129), TMEM129ΔRING (ΔRING) or an empty vector (ctrl). (f) Same cells as in Fig. 3e, now analysed by immunoblotting for the indicated proteins. TMEM129-FLAG retained its ability to enhance HLA class I downregulation (Supplementary Fig. 10a).
Figure 4. TMEM129 is part of the US11 dislocation complex.
Strep-tag II- and HA-tagged US11 was immunoprecipitated using StrepTactin beads from 1.0% digitonin lysates of US11-negative (lane 1) and positive (lanes 2–6) U937 eGFP-Myc-HLA-A2 cells. The US11-expressing cells were co-expressing a control shRNA (shCtrl, lane 2), an shRNA targeting TMEM129 (shT129, lane 3), an empty vector (ctrl, lane 4), TMEM129 (T129, lane 5) or TMEM129ΔRING (ΔRING, lane 6). Immunoprecipitated proteins were eluted using d-Desthiobiotin, after which immunoblot analysis was performed for proteins indicated. The right panels (lanes 7–12) indicate loading controls to analyse input of the indicated proteins prior immunoprecipitation. The Strep-tag II- and HA-tagged US11 retained its ability to downregulate HLA class I molecules (Supplementary Fig. 10b). The HRD1 antibody could not detect HRD1 in digitonin cell lysates, indicated by the asterisk. The double asterisk indicates an additional unspecified truncated form of TMEM129ΔRING.
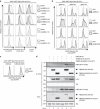
Figure 5. UBE2J2 is essential for US11-mediated HLA class I downregulation.
(a) UBE2J2 depletion by shRNAs induces rescue of HLA class I in US11-expressing cells. Two individual UBE2J2- and UBE2J1-targeting shRNAs (grey-lined histograms), two individual UBE2J2-targeting shRNAs together with UBE2J2 cDNA (black-lined histograms), or one control shRNA (dashed histograms) were introduced in U937 eGFP-Myc-HLA-A2 US11 cells. The flow cytometry analysis of endogenous surface HLA-A3, and surface (Myc) and total (eGFP) eGFP-Myc-HLA-A2 expression was performed at 7 dpi. (b) CRISPR/Cas-mediated knockout of UBE2J2 and UBE2K induce potent rescue of HLA class I. Total (eGFP) expression of eGFP-Myc-HLA-A2 were assessed by flow cytometry in U937 eGFP-Myc-HLA-A2 US11 control cells (grey histogram) and cells knocked out for either UBE2J2 (black histograms, upper panels), UBE2K (black histograms; second panels), UBE2J1 (black histograms, third panels) or TMEM129 (black histograms, lower panels) using three individual CRISPR gRNAs. (c) Dominant-negative UBE2J2 (C94S) causes rescue of HLA class I. Total (eGFP) expression of eGFP-Myc-HLA-A2 was assessed by flow cytometry in U937 eGFP-Myc-HLA-A2 US11 control cells (grey histogram) and cells expressing either wild-type UBE2J2 (black histogram, left panel) or dominant-negative UBE2J2-C94S (black histogram, right panel). (d) UBE2J2 associates with TMEM129. Strep-tag II- and FLAG-tagged TMEM129 was immunoprecipitated using StrepTactin beads from 1.0% digitonin lysates of U937 eGFP-Myc-HLA-A2 US11 cells expressing indicated constructs. The Strep-tag II- and FLAG-tagged TMEM129 retained its ability to enhance HLA class I downregulation (Supplementary Fig. 10c, left panels). Immunoprecipitated proteins were eluted using d-Desthiobiotin, after which immunoblot analysis was performed for the proteins indicated.
Figure 6. TMEM129 is present in ERAD complexes in the absence of US11.
(a) C-terminally Strep-tag-II- and FLAG-tagged TMEM129 or TMEM129ΔRING were immunoprecipitated using StrepTactin beads from 1.0% digitonin lysates of US11-negative (lanes 1–3) and US11-positive (lanes 4–6) U937 eGFP-Myc-HLA-A2 cells. The cells were co-expressing a control vector (ctrl, lanes 1 and 4), TMEM129-FLAG-ST2 (T129, lanes 2 and 5), or TMEM129ΔRING-FLAG-ST2 (ΔRING, lanes 3 and 6). Immunoprecipitated proteins were eluted using d-Desthiobiotin, after which immunoblot analysis was performed for the proteins indicated. The right panels (lanes 7–12) indicate loading controls to analyse input of the indicated proteins before immunoprecipitation. The C-terminally Strep-tag-II- and FLAG-tagged TMEM129 construct retained its ability to enhance US11-mediated HLA class I downregulation (Supplementary Fig. 10c, left panels), whereas the C-terminally tagged TMEM129ΔRING retained its dominant-negative phenotype (Supplementary Fig. 10c, right panels). The HRD1 antibody could not detect HRD1 in digitonin cell lysates, indicated by the asterisk. (b) C-terminally FLAG-tagged TMEM129 and HRD1 were immunoprecipitated using FLAG-M2-coupled beads from 1.0% digitonin lysates of US11-negative (lanes 1–3) and US11-positive (lanes 4–6) U937 eGFP-Myc-HLA-A2 cells. Immunoprecipitated proteins were eluted using FLAG peptides, after which immunoblot analysis was performed for the proteins indicated. (c) Schematic overview of the US11 dislocation complex. The depicted location of the proteins is not accurate.
Similar articles
- TMEM129 is a Derlin-1 associated ERAD E3 ligase essential for virus-induced degradation of MHC-I.
van den Boomen DJ, Timms RT, Grice GL, Stagg HR, Skødt K, Dougan G, Nathan JA, Lehner PJ. van den Boomen DJ, et al. Proc Natl Acad Sci U S A. 2014 Aug 5;111(31):11425-30. doi: 10.1073/pnas.1409099111. Epub 2014 Jul 16. Proc Natl Acad Sci U S A. 2014. PMID: 25030448 Free PMC article. - Identifying the ERAD ubiquitin E3 ligases for viral and cellular targeting of MHC class I.
van den Boomen DJ, Lehner PJ. van den Boomen DJ, et al. Mol Immunol. 2015 Dec;68(2 Pt A):106-11. doi: 10.1016/j.molimm.2015.07.005. Epub 2015 Jul 22. Mol Immunol. 2015. PMID: 26210183 Free PMC article. Review. - Human cytomegalovirus evades antibody-mediated immunity through endoplasmic reticulum-associated degradation of the FcRn receptor.
Liu X, Palaniyandi S, Zhu I, Tang J, Li W, Wu X, Ochsner SP, Pauza CD, Cohen JI, Zhu X. Liu X, et al. Nat Commun. 2019 Jul 9;10(1):3020. doi: 10.1038/s41467-019-10865-y. Nat Commun. 2019. PMID: 31289263 Free PMC article. - The E3 Ubiquitin Ligase TMEM129 Is a Tri-Spanning Transmembrane Protein.
van de Weijer ML, van Muijlwijk GH, Visser LJ, Costa AI, Wiertz EJ, Lebbink RJ. van de Weijer ML, et al. Viruses. 2016 Nov 15;8(11):309. doi: 10.3390/v8110309. Viruses. 2016. PMID: 27854284 Free PMC article. - The role of p97/Cdc48p in endoplasmic reticulum-associated degradation: from the immune system to yeast.
Bar-Nun S. Bar-Nun S. Curr Top Microbiol Immunol. 2005;300:95-125. doi: 10.1007/3-540-28007-3_5. Curr Top Microbiol Immunol. 2005. PMID: 16573238 Review.
Cited by
- Alternative Antigen Processing for MHC Class I: Multiple Roads Lead to Rome.
Oliveira CC, van Hall T. Oliveira CC, et al. Front Immunol. 2015 Jun 5;6:298. doi: 10.3389/fimmu.2015.00298. eCollection 2015. Front Immunol. 2015. PMID: 26097483 Free PMC article. Review. - Molecular basis determining species specificity for TLR2 inhibition by staphylococcal superantigen-like protein 3 (SSL3).
Koymans KJ, Feitsma LJ, Bisschop A, Huizinga EG, van Strijp JAG, de Haas CJC, McCarthy AJ. Koymans KJ, et al. Vet Res. 2018 Nov 28;49(1):115. doi: 10.1186/s13567-018-0609-8. Vet Res. 2018. PMID: 30486901 Free PMC article. - Mechanisms of substrate processing during ER-associated protein degradation.
Christianson JC, Jarosch E, Sommer T. Christianson JC, et al. Nat Rev Mol Cell Biol. 2023 Nov;24(11):777-796. doi: 10.1038/s41580-023-00633-8. Epub 2023 Aug 1. Nat Rev Mol Cell Biol. 2023. PMID: 37528230 Review. - Molecular mechanism of ER stress-induced pre-emptive quality control involving association of the translocon, Derlin-1, and HRD1.
Kadowaki H, Satrimafitrah P, Takami Y, Nishitoh H. Kadowaki H, et al. Sci Rep. 2018 May 9;8(1):7317. doi: 10.1038/s41598-018-25724-x. Sci Rep. 2018. PMID: 29743537 Free PMC article. - Knockout of cGAS and STING Rescues Virus Infection of Plasmid DNA-Transfected Cells.
Langereis MA, Rabouw HH, Holwerda M, Visser LJ, van Kuppeveld FJ. Langereis MA, et al. J Virol. 2015 Nov;89(21):11169-73. doi: 10.1128/JVI.01781-15. Epub 2015 Aug 26. J Virol. 2015. PMID: 26311870 Free PMC article.
References
- Kim Y. E., Hipp M. S., Bracher A., Hayer-Hartl M. & Hartl F. U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355 (2013). - PubMed
- Hampton R. Y. ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 14, 476–482 (2002). - PubMed
- Amm I., Sommer T. & Wolf D. H. Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim. Biophys. Acta 1843, 182–196 (2014). - PubMed
- Merulla J., Fasana E., Soldà T. & Molinari M. Specificity and regulation of the endoplasmic reticulum-associated degradation machinery. Traffic 14, 767–777 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM080783/GM/NIGMS NIH HHS/United States
- U01 CA168370/CA/NCI NIH HHS/United States
- R01 GM80783/GM/NIGMS NIH HHS/United States
- 1U01CA168370-01/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials