Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression - PubMed (original) (raw)
. 2014 Jul 31;511(7511):601-5.
doi: 10.1038/nature13554. Epub 2014 Jul 9.
Steven E Bosinger 2, Jacob D Estes 3, Richard T R Zhu 4, Gregory K Tharp 2, Eli Boritz 4, Doron Levin 5, Sathi Wijeyesinghe 4, Krystelle Nganou Makamdop 4, Gregory Q del Prete 3, Brenna J Hill 4, J Katherina Timmer 4, Emma Reiss 4, Ganit Yarden 5, Samuel Darko 4, Eduardo Contijoch 4, John Paul Todd 6, Guido Silvestri 7, Martha Nason 8, Robert B Norgren Jr 9, Brandon F Keele 3, Srinivas Rao 6, Jerome A Langer 10, Jeffrey D Lifson 3, Gideon Schreiber 5, Daniel C Douek 4
Affiliations
- PMID: 25043006
- PMCID: PMC4418221
- DOI: 10.1038/nature13554
Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression
Netanya G Sandler et al. Nature. 2014.
Abstract
Inflammation in HIV infection is predictive of non-AIDS morbidity and death, higher set point plasma virus load and virus acquisition; thus, therapeutic agents are in development to reduce its causes and consequences. However, inflammation may simultaneously confer both detrimental and beneficial effects. This dichotomy is particularly applicable to type I interferons (IFN-I) which, while contributing to innate control of infection, also provide target cells for the virus during acute infection, impair CD4 T-cell recovery, and are associated with disease progression. Here we manipulated IFN-I signalling in rhesus macaques (Macaca mulatta) during simian immunodeficiency virus (SIV) transmission and acute infection with two complementary in vivo interventions. We show that blockade of the IFN-I receptor caused reduced antiviral gene expression, increased SIV reservoir size and accelerated CD4 T-cell depletion with progression to AIDS despite decreased T-cell activation. In contrast, IFN-α2a administration initially upregulated expression of antiviral genes and prevented systemic infection. However, continued IFN-α2a treatment induced IFN-I desensitization and decreased antiviral gene expression, enabling infection with increased SIV reservoir size and accelerated CD4 T-cell loss. Thus, the timing of IFN-induced innate responses in acute SIV infection profoundly affects overall disease course and outweighs the detrimental consequences of increased immune activation. Yet, the clinical consequences of manipulation of IFN signalling are difficult to predict in vivo and therapeutic interventions in human studies should be approached with caution.
Conflict of interest statement
The authors declare competing financial interests: details are available in the online version of the paper. Readers are welcome to comment on the online version of the paper.
Figures
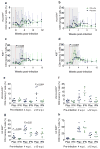
Extended Data Figure 1. Dose escalation study for IFN-1ant and experimental schema
a–d, Effects of three times weekly IFN-1ant dosing on the frequency of CD4 T cells (a), CCR5+ CD4 T cells (b), CCR5+ CD8 T cells (c) and Ki67+ CD8 T cells (d) in 2 rhesus macaques. Dose was 50 μg in week 1, 200 μg in week 2, 500 μg in week 3 and 800 μg in week 4. Vertical dotted lines indicate the days a new dose was started. Black lines connect time points 4 days after the first dose. Grey shading indicates treatment period. e, Six macaques received 4 weeks of IFN-1ant intramuscularly starting at day 0 and were challenged intrarectally with 1 ml of a 1:25 dilution of SIVMAC251 (stock concentration 3 ×108 SIV RNA copies ml−1) at day 0 and followed until developing end-stage AIDS. Nine macaques were treated with 4 weeks of placebo saline intramuscularly starting at day 0 and challenged intrarectally with SIVMAC251 at day 0 and followed. Six macaques were injected weekly with IFN-α2a starting 1 week before the first challenge and through 4 w.p.i. Macaques required 2, 3 or 5 challenges to acquire systemic infection. Thus, macaques received 6, 7 or 9 doses of IFN-α2a. Macaques were necropsied at 12 w.p.i. per protocol.
Extended Data Figure 2. Effects of IFN-1ant on IFN-stimulated genes and virus burden
a, b, MX1 (a) and OAS2 (b) expression by qRT–PCR during acute SIV infection in IFN-1ant (red, n =6) and placebo (blue, n =9) macaques. P values were calculated by Mann–Whitney U test. c, ISGs in PBMCs in IFN-1ant and placebo macaques. P values represent the comparison between IFN-1ant (n =6) and placebo (n =9) macaque FPKMs at 7 d.p.i. d, e, SAMHD1 (d) and APOBEC3G (e) expression in the lymph nodes in IFN-1ant (n =6) and placebo (n =9) macaques. P values were calculated by Mann–Whitney U test. f, g, Plasma SIV RNA levels at 12 w.p.i. (f) or at peak (g) stratified by the day that MX1 or OAS2 expression peaked in PBMCs in IFN-1ant (n =6) and placebo (n =9) macaques. VL, viral load. P values were calculated by Mann–Whitney U test. h, SIV gag levels in PBMCs stratified by the day that MX1 or OAS2 expression peaked in PBMCs in IFN-1ant (n =6) and placebo (n =6) macaques. P values were calculated by Mann–Whitney U test. For all panels, IFN-1ant-treated macaques are represented in red, placebo-treated macaques in blue.
Extended Data Figure 3. Effects of IFN-1ant on CD4 T cells and on immune activation
a, b, CD4/CD8 T-cell ratio in peripheral blood (a) and lymph node (LN) (b) in IFN-1ant (Ant, n =6) and placebo (Plac, n =9) macaques. Shading indicates treatment period. Error bars indicate range. Red vertical line indicates day 0 of systemic SIV infection. For all panels, horizontal bars indicate median values, and P values at different time points within treatment groups were calculated by Wilcoxon matched pairs signed rank test and between groups by Mann–Whitney U test. c–f, T-cell activation in lymph nodes (c–f) in CD4 (c, d) and CD8 (e, f) T cells as represented by the frequency of Ki67+ (c, e) or HLA-DR+ (d, f) cells in IFN-1ant (n =6) and placebo (n =9) macaques. g, Frequency of circulating CD16+ or CD56+CD3−CD14− NK cells in IFN-1ant (n =6) and placebo (n =9) macaques. h, Frequency of circulating CD16+ NK cells in IFN-1ant (n =6) and placebo (n =9) macaques. i, Frequency of circulating CD56+ NK cells in IFN-1ant (n =6) and placebo (n =9) macaques. For all panels, IFN-1ant-treated macaques are represented in red, placebo-treated macaques in blue.
Extended Data Figure 4. IFN-1ant alters innate and adaptive immune signalling
a, Selected pathways significantly affected by IFN-I blockade. P values were calculated by Fisher’s exact test with the Benjamini–Hochberg multiple testing correction. b, Expression of genes involved in pattern recognition receptor signalling of IFN-1ant-treated macaques (n =6) compared to placebo (n =9) at 7 d.p.i. Upregulation compared to pre-infection is represented by red, no change by white, downregulation by blue. P values represent the comparison between IFN-1ant and placebo macaques at 7 d.p.i. c, Selected genes in pattern recognition receptor signalling pathways. Upregulation at 7 d.p.i. is represented by red, downregulation by green.
Extended Data Figure 5. Effects of IFN-1ant on T-cell function and phenotype
a–e, SIV-specific responses in peripheral blood at 4 and >12 w.p.i. in IFN-1ant (Ant, n =6) and placebo (Plac, n =6) macaques by frequency of IFN-γ+(a), TNF+ (b), perforin+ (c), granzyme B+ (d) and CD107+ (e) CD8 T cells. T-cell exhaustion in peripheral blood and lymph nodes (LN) at >16 w.p.i. based on frequency of PD-1+ CD4 (f) and CD8 (h) T cells and ICOS+ (g) CD8 T cells. For all panels, P values at different time points within treatment groups were calculated by Wilcoxon matched pairs signed rank test and between groups by Mann–Whitney U test. IFN-1ant-treated macaques are represented in red, placebo-treated macaques in blue.
Extended Data Figure 6. IFN-α2a treatment transiently induces ISGs and subsequently induces the IFN-repressor FOXO3a but does not induce neutralizing anti-IFN antibodies
a–d, MX1 (a, c) and OAS2 (b, d) expression during the duration of IFN-α2a treatment in the IFN-α2a group alone (a, b) and during infection in the IFN-α2a (n =6) and placebo (n =9) groups (c, d). P values were calculated by Wilcoxon matched pairs signed rank test. e, Percentage of in vitro IFN antiviral activity inhibited by plasma from IFN-α2a (n =6) and placebo (n =3) macaques. f, Expression of FOXO3a and FOXO3a-bound genes in SIV-uninfected macaques (n =3) treated with 21 days of IFN-α2a. Large circles indicate statistically significant (P<0.05) changes from pre-IFN-α2a treatment calculated by Wilcoxon matched pairs signed rank test. Small circles indicate no statistically significant change from pre-IFN-α2a treatment. g, Expression of IFN-α-regulatory genes in IFN-α2a (n =6) and placebo (n =9) macaques. P values represent the comparison between FPKMs of IFN-α2a (n =6) and placebo (n =9) macaques at 7 d.p.i. h, Expression of FOXO3a-bound genes in IFN-α2a (green, n =6) and placebo (blue, n =9) macaques at 7 d.p.i.
Extended Data Figure 7. Effects of IFN-α2a on IFN-stimulated and antiviral genes
a, ISGs in PBMCs in IFN-α2a (n =6) and placebo (n =9) macaques. Red indicates upregulation, yellow indicates no change and blue indicates downregulation relative to pre-infection. b, Expression of ISGs in macaques treated with IFN-α2a (n =6) or placebo (n =9). P values indicate differentially expressed genes at 10 d.p.i. c–h, Expression of TRIM22 (c, d), MX2 (e, f) and IRF7 (g, h) in SIV-uninfected macaques (n =3) treated with weekly IFN-α2a for 3 weeks in PBMCs (c, e, g) and lymph nodes and rectum (d, f, h). Day 0 reflects baseline. Numbers indicate days since first IFN-α2a administration. Error bars indicate range. P values were calculated by Wilcoxon matched pairs signed rank test.
Extended Data Figure 8. Effects of IFN-α2a on SIV control
a, Number of transmitted/founder (T/F) variants in placebo (n =9), IFN-1ant (n =6) and IFN-α2a (n =6) macaques. P value was calculated by Mann–Whitney U test. b, Antiviral protein production in lymph nodes (LN) by immunohistochemistry at 4 w.p.i. in IFN-α2a (n =6) and placebo (n =6) macaques. P value was calculated by Mann–Whitney U test. c, CD56+ NK-cell frequency on the day of challenge stratified by whether the macaque resisted or was susceptible to systemic infection that day. Each IFN-α2a macaque (n =6) is indicated by a different colour. Circles indicate that the macaque was resistant to infection with the next challenge and triangles indicate that the macaque was susceptible to infection with the next challenge. P value was calculated by Mann–Whitney U test. d, Correlation between the number of challenges required to achieve systemic infection and rectal CD16+ NK-cell frequency in each macaque (n =6) at 4 w.p.i. r indicates the Spearman’s rank correlation coefficient. P value indicates the significance of the correlation. e, Plasma SIV RNA levels in macaques treated with IFN-α2a (n =6) or placebo (n =9) saline. Shading reflects treatment period. Red vertical line indicates day 0 of systemic SIV infection. f–i, Frequency of IFN-γ+ (f), TNF+ (g), granzyme B+ (h) and perforin+ (i) CD8 T cells at 4 and ≥12 w.p.i. in IFN-α2a (n =6) and placebo (n =6) macaques. j, Frequency of circulating CD16+CD56− NK cells in IFN-α2a (n =6) and placebo (n =9) macaques. P values at different time points within treatment groups were calculated by Wilcoxon matched pairs signed rank test and between groups by Mann–Whitney U test.
Extended Data Figure 9. Effects of IFN-α2a on T-cell activation
a–h, Frequency of peripheral blood (a–d) and lymph node (LN) (e–h) CD4 (a, c, e, g) and CD8 (b, d, f, h) memory T cells expressing HLA-DR (a, b, e, f) or Ki67 (c, d, g, h) in IFN-α2a (IFN, n =6) and placebo (Plac, n =9) macaques. Shading indicates treatment period. Error bars indicate range. a–d, Red vertical line indicates day 0 of systemic SIV infection. P values represent the comparison between groups of the AUC (0–4 w.p.i.). e–h, Horizontal bars indicate median values. P values were calculated by Mann–Whitney U test.
Extended Data Figure 10. Effects of IFN-α2a on gene expression
a, Selected pathways significantly affected by IFN-α2a treatment. P values were calculated by Fisher’s exact test with the Benjamini–Hochberg multiple testing correction. b, Expression of genes downstream of IL-6 signalling. Upregulation relative to before IFN-α2a or placebo treatment and SIV infection is represented by red, no change by white, downregulation by blue. P values represent the comparison between IFN-α2a (n =6) and placebo (n =9) macaques at 7 d.p.i. c, Selected genes in apoptosis signalling pathways. Significant upregulation at 7 d.p.i. is represented by red, downregulation by green.
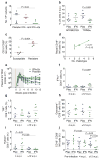
Figure 1. IFN-1ant suppresses early antiviral responses
a, Expression of ISGs in macaques treated with IFN-1ant (n =6) or placebo saline (n =9) 7 days after SIV infection. FPKM (log-transformed fragments per kilobase of transcript per million fragments sequenced) reflects the relative abundance of transcripts. P values indicate differentially expressed genes at 7 d.p.i. b, Expression assessed by RNA sequencing (RNA-seq) of antiviral genes APOBEC3G, MX2 and those that code for cGAS and tetherin in PBMCs before and 7 days after SIV infection in macaques that received IFN-1ant (Ant, n =6) or placebo (Plac, n =9) injections. Error bars indicate range. P values were calculated by Mann–Whitney U test. c, APOBEC3G, TRIM5α and MX2 protein expression by immunohistochemistry of lymph nodes (LNs) at 4 w.p.i. in placebo (n =6) and IFN-1ant (n =6) macaques. Horizontal bars represent median values. P values were calculated by Mann–Whitney U test. The left panels are representative images of APOBEC3G staining from each group. d, Expression of genes involved in pattern recognition receptor signalling of IFN-1ant-treated (n =6) macaques compared to placebo (n =9). P values represent the differential expression between IFN-1ant and placebo macaques at 7 d.p.i. For all panels, IFN-1ant-treated macaques are represented in red, placebo-treated macaques in blue.
Figure 2. IFN-1ant accelerates disease progression in SIV-infected rhesus macaques
a, Plasma SIV RNA levels during acute and chronic SIV infection in macaques treated with IFN-1ant (n =6) or placebo saline (n =9). *P <0.05. Shading indicates treatment period. _P_ value represents the comparison between groups of the areas under the curve (AUC) (0–4 w.p.i.). **b**, SIV RNA-containing cells in the lymph nodes by _in situ_ hybridization at 4 and 12 w.p.i. in IFN-1ant (Ant, _n_ =6) and placebo (Plac, _n_ =6) macaques. Horizontal bars represent median values. _P_ value was calculated by Mann–Whitney _U_ test. **c**, Frequency of CD4 T cells in peripheral blood during acute and chronic SIV infection in macaques treated with IFN-1ant (_n_ =6) or placebo saline (_n_ =9). Error bars indicate range. Red vertical line indicates day 0 of systemic SIV infection. Shading indicates treatment period. _P_ value represents the comparison between groups of the AUC (12–32 w.p.i.). **d**, Frequency of CD4 T cells in lymph nodes before SIV infection and at 4 or >12 w.p.i. for IFN-1ant (Ant, n =6) and placebo (Plac, n =9) macaques. Horizontal bars represent median values. P values at different time points within treatment groups were calculated by Wilcoxon matched pairs signed rank test and between groups by Mann–Whitney U test. e, Frequency of CCR5+ memory (CD28+CD95+ or CD28−CD95+/−) CD4 T cells in peripheral blood in macaques treated with IFN-1ant (n =6) or placebo saline (n =9). Error bars indicate range. Red vertical line indicates day 0 of systemic SIV infection. Shading indicates treatment period. P values represent the comparison between groups of the AUC (0–12 w.p.i. and 4–12 w.p.i.). f, Frequency of CCR5+ memory CD4 T cells in lymph nodes in macaques treated with IFN-1ant (n =6) or placebo saline (n =9). Horizontal bars represent median values. P values at different time points within treatment groups were calculated by Wilcoxon matched pairs signed rank test and between groups by Mann–Whitney U test. g, Kaplan–Meier survival curve comparing macaques treated with IFN-1ant (n =6) to macaques that received placebo (n =9). P value indicates the significance by log rank (Mantel–Cox) test for survival by 32 w.p.i. For all panels, IFN-1ant-treated macaques are represented in red, placebo-treated macaques in blue.
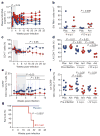
Figure 3. IFN-α2a treatment transiently prevents systemic infection but results in an IFN-tolerant state
a, Kaplan–Meier survival curve comparing the number of SIVMAC251 rectal challenges required to achieve systemic infection in macaques treated with IFN-α2a (n =6) or placebo saline (n =9). P value indicates the significance by logrank (Mantel–Cox) test of the number of challenges required for systemic infection, between 1 and 5 challenges. b, Correlation between the number of challenges needed to achieve systemic infection and the number of transmitted/founder (T/F) variants in IFN-α2a (green, n =6), IFN-1ant (red, n =6) and placebo (blue, n =9) macaques. P value indicates the significance of the correlation between the number of challenges and the number of T/F variants in all groups. r indicates the Spearman’s rank correlation coefficient. c–f, Expression of antiviral mediators in PBMCs in IFN-α2a-treated (IFN, n =6) macaques compared to placebo (Plac, n =9) at 10 d.p.i. Error bars indicate range. P values represent the comparison of FPKMs between IFN-α2a and placebo at 10 d.p.i. by Mann–Whitney U test. g, Expression profile of FOXO3a, a negative regulator of type I IFN signalling. P value represents the comparison of FOXO3a FPKM between IFN-α2a (n =6) and placebo (n =9) macaques at 7 d.p.i. h, Gene-set enrichment analysis in IFN-α2a (n =6) and placebo (n =9) macaques of genes previously demonstrated to be overexpressed in _FOXO3_−/− macrophages. The line plot indicates the running-sum of the enrichment score; the leading edge is indicated in magenta. The relative positions of all genes within the ranked data set are shown in the stick plot below the x axis. P value indicates statistical significance of the enrichment score, reflecting lower cumulative ranking of FOXO3a targets in IFN-α2a-treated macaques compared to placebo at 7 d.p.i. For all panels, IFN-α2a-treated macaques are represented in green, placebo-treated macaques in blue.
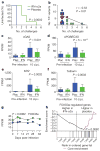
Figure 4. IFN-α2a accelerates disease progression
a, PBMC-associated SIV gag DNA at 10, 14 and 28 d.p.i. in IFN-α2a macaques (IFN, n =6) and placebo (Plac, n =6) macaques. LLQ indicates lower limit of quantification. Horizontal bars represent median values. P values were calculated by Mann–Whitney U test. b, Expression of differentially expressed genes involved in pattern recognition receptor signalling. P values represent the comparison between FPKMs of IFN-α2a (n =6) and placebo (n =9) macaques at 7 d.p.i. c, Frequency of CD4 T cells in peripheral blood during acute and early SIV infection in IFN-α2a (n =6) and placebo (n =9) macaques. Error bars indicate range. Shading indicates treatment period. Red vertical line indicates day 0 of systemic SIV infection. d, CD4/CD8 T-cell ratio in peripheral blood during acute and early SIV infection in IFN-α2a (n =6) and placebo (n =9) macaques. Error bars indicate range. Shading indicates treatment period. Red vertical line indicates day 0 of systemic SIV infection. P value represents the comparison between groups of the AUC (0–12 w.p.i.). e, Frequency of CCR5+ memory CD4 T cells in peripheral blood in IFN-α2a (n =6) and placebo (n =9) macaques. Error bars indicate range. Shading indicates treatment period. Red vertical line indicates day 0 of systemic SIV infection. P values represent the comparison between groups of the AUC (0–4 and 0–12 w.p.i.). f, Frequency of CCR5+ memory CD4 T cells in lymph nodes in IFN-α2a (n =6) and placebo (n =9) macaques. Horizontal bars represent median values. P values were calculated by Mann–Whitney U test. For all panels, IFN-α2a-treated macaques are represented in green, placebo-treated macaques in blue.
Comment in
- HIV: The mixed blessing of interferon.
Telenti A. Telenti A. Nature. 2014 Jul 31;511(7511):537-8. doi: 10.1038/nature13517. Epub 2014 Jul 9. Nature. 2014. PMID: 25043060 No abstract available.
Similar articles
- Short-Term Pegylated Interferon α2a Treatment Does Not Significantly Reduce the Viral Reservoir of Simian Immunodeficiency Virus-Infected, Antiretroviral Therapy-Treated Rhesus Macaques.
Palesch D, Bosinger SE, Mavigner M, Billingsley JM, Mattingly C, Carnathan DG, Paiardini M, Chahroudi A, Vanderford TH, Silvestri G. Palesch D, et al. J Virol. 2018 Jun 29;92(14):e00279-18. doi: 10.1128/JVI.00279-18. Print 2018 Jul 15. J Virol. 2018. PMID: 29720521 Free PMC article. - Reduced Chronic Lymphocyte Activation following Interferon Alpha Blockade during the Acute Phase of Simian Immunodeficiency Virus Infection in Rhesus Macaques.
Carnathan D, Lawson B, Yu J, Patel K, Billingsley JM, Tharp GK, Delmas OM, Dawoud R, Wilkinson P, Nicolette C, Cameron MJ, Sekaly RP, Bosinger SE, Silvestri G, Vanderford TH. Carnathan D, et al. J Virol. 2018 Apr 13;92(9):e01760-17. doi: 10.1128/JVI.01760-17. Print 2018 May 1. J Virol. 2018. PMID: 29467313 Free PMC article. - Elite Control, Gut CD4 T Cell Sparing, and Enhanced Mucosal T Cell Responses in Macaca nemestrina Infected by a Simian Immunodeficiency Virus Lacking a gp41 Trafficking Motif.
Breed MW, Elser SE, Torben W, Jordan AP, Aye PP, Midkiff C, Schiro F, Sugimoto C, Alvarez-Hernandez X, Blair RV, Somasunderam A, Utay NS, Kuroda MJ, Pahar B, Wiseman RW, O'Connor DH, LaBranche CC, Montefiori DC, Marsh M, Li Y, Piatak M Jr, Lifson JD, Keele BF, Fultz PN, Lackner AA, Hoxie JA. Breed MW, et al. J Virol. 2015 Oct;89(20):10156-75. doi: 10.1128/JVI.01134-15. Epub 2015 Jul 29. J Virol. 2015. PMID: 26223646 Free PMC article. - Manipulating the Interferon Signaling Pathway: Implications for HIV Infection.
Nganou-Makamdop K, Douek DC. Nganou-Makamdop K, et al. Virol Sin. 2019 Apr;34(2):192-196. doi: 10.1007/s12250-019-00085-5. Epub 2019 Feb 14. Virol Sin. 2019. PMID: 30762199 Free PMC article. Review.
Cited by
- The importance of IFNα2A (Roferon-A) in HSV-1 latency and T cell exhaustion in ocularly infected mice.
Wang S, Jaggi U, Katsumata M, Ghiasi H. Wang S, et al. PLoS Pathog. 2024 Oct 1;20(10):e1012612. doi: 10.1371/journal.ppat.1012612. eCollection 2024 Oct. PLoS Pathog. 2024. PMID: 39352890 Free PMC article. - Intravenous Bacillus Calmette-Guérin (BCG) Induces a More Potent Airway and Lung Immune Response than Intradermal BCG in Simian Immunodeficiency Virus-infected Macaques.
Jauro S, Larson EC, Gleim JL, Wahlberg BM, Rodgers MA, Chehab JC, Lopez-Velazques AE, Ameel CL, Tomko JA, Sakal JL, DeMarco T, Borish HJ, Maiello P, Potter EL, Roederer M, Ling Lin P, Flynn JL, Scanga CA. Jauro S, et al. J Immunol. 2024 Nov 1;213(9):1358-1370. doi: 10.4049/jimmunol.2400417. J Immunol. 2024. PMID: 39311665 - Microglia and macrophages alterations in the CNS during acute SIV infection: A single-cell analysis in rhesus macaques.
Xu X, Niu M, Lamberty BG, Emanuel K, Ramachandran S, Trease AJ, Tabassum M, Lifson JD, Fox HS. Xu X, et al. PLoS Pathog. 2024 Sep 16;20(9):e1012168. doi: 10.1371/journal.ppat.1012168. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39283947 Free PMC article. - A cGAS-mediated type I interferon response in human CD4+ T cells depends on productive infection and is conserved over HIV types and strains.
Janevska M, Cammaert T, Naessens E, Verhasselt B. Janevska M, et al. J Virol. 2024 Oct 22;98(10):e0087724. doi: 10.1128/jvi.00877-24. Epub 2024 Sep 13. J Virol. 2024. PMID: 39269176 - Immune perturbation following SHIV infection is greater in newborn macaques than in infants.
Shapiro MB, Ordonez T, Pandey S, Mahyari E, Onwuzu K, Reed J, Sidener H, Smedley J, Colgin LM, Johnson A, Lewis AD, Bimber B, Sacha JB, Hessell AJ, Haigwood NL. Shapiro MB, et al. JCI Insight. 2024 Aug 27;9(19):e144448. doi: 10.1172/jci.insight.144448. JCI Insight. 2024. PMID: 39190496 Free PMC article.
References
- Hunt PW, et al. Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J Infect Dis. 2014 Apr 21; http://dx.doi.org/10.1093/infdis/jiu238. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- ZIA AI005034-13/ImNIH/Intramural NIH HHS/United States
- AI-076174/AI/NIAID NIH HHS/United States
- HHSN261200800001C/CA/NCI NIH HHS/United States
- R24 RR017444/RR/NCRR NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- P01 AI076174/AI/NIAID NIH HHS/United States
- P51 OD011132/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials