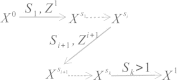A new method for modeling coalescent processes with recombination - PubMed (original) (raw)
A new method for modeling coalescent processes with recombination
Ying Wang et al. BMC Bioinformatics. 2014.
Abstract
Background: Recombination plays an important role in the maintenance of genetic diversity in many types of organisms, especially diploid eukaryotes. Recombination can be studied and used to map diseases. However, recombination adds a great deal of complexity to the genetic information. This renders estimation of evolutionary parameters more difficult. After the coalescent process was formulated, models capable of describing recombination using graphs, such as ancestral recombination graphs (ARG) were also developed. There are two typical models based on which to simulate ARG: back-in-time model such as ms and spatial model including Wiuf&Hein's, SMC, SMC', and MaCS.
Results: In this study, a new method of modeling coalescence with recombination, Spatial Coalescent simulator (SC), was developed, which considerably improved the algorithm described by Wiuf and Hein. The present algorithm constructs ARG spatially along the sequence, but it does not produce any redundant branches which are inevitable in Wiuf and Hein's algorithm. Interestingly, the distribution of ARG generated by the present new algorithm is identical to that generated by a typical back-in-time model adopted by ms, an algorithm commonly used to model coalescence. It is here demonstrated that the existing approximate methods such as the sequentially Markov coalescent (SMC), a related method called SMC', and Markovian coalescent simulator (MaCS) can be viewed as special cases of the present method. Using simulation analysis, the time to the most common ancestor (TMRCA) in the local trees of ARGs generated by the present algorithm was found to be closer to that produced by ms than time produced by MaCS. Sample-consistent ARGs can be generated using the present method. This may significantly reduce the computational burden.
Conclusion: In summary, the present method and algorithm may facilitate the estimation and description of recombination in population genomics and evolutionary biology.
Figures
Figure 1
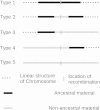
Five types of recombination in the history of a population.
Figure 2
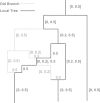
An example of current ARG. The graph displays the [0, 0.5] part of an ARG, the black lines represents branches constituting the current local tree, the gray lines represent all old branches, the numbers in brackets display intervals which denote the ancestral materials carried by nearby branches, the numbers without brackets denote recombination rates occur in the underlying nodes.
Figure 3
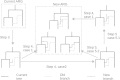
Generation of ARG along sequence. X s denotes [0, s] segments of the ARG, S i denotes the i th type 1 recombination breakpoints and Z i denotes the branches added to X Si_-_1. X Si is the collection of Z i and X Si_-_1.
Figure 4
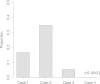
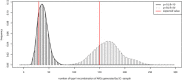
Comparison of differences in the mean and variance of the first 100 local trees’ height between SC and Macs using ms as a control. Boxplot with 75% quantile and 25% quantile as top border and the bottom border, respectively.
Figure 5
Ratio of strictly sample-consistent ARGs to all ARGs. ARGs are not considered strictly sample-consistent unless they are both sample-consistent and the number of type 1 recombination of that ARG is within 10% of estimate using 100,000 simulations in 4 different cases. Case 1: ρ = 10, μ = 10 with _SC_-sample. Case 2: ρ = 50, μ = 50 with _SC_-sample. Case 3: ρ = 16.9,μ = 7.5 with _SC_-sample, which employ the mutation rate and recombination rate in human. Case 4: ρ = 10, μ = 10 with SC.
Figure 6
Distribution of number of type1 recombination in ARGs generated with SC - sample. The red vertical line indicates the expected number of type 1 recombination in a particular scenario**.**
Figure 7
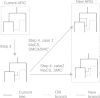
Updated steps of SC to generate new ARG from current ARG. The same step numbers and case numbers as in the methods section are used here. Step 3 is a new type 1 recombination created on the current ARG. In step 4, the new branch coalesces into an old branch or a branch on current tree. If the new branch coalesces into the current tree, a new ARG has been constructed. If the new branch coalesces into an old branch, then there are two cases, case 5.1 and case 5.2. In case 5.2, a new ARG is generated. In case 5.1, a new branch is generated which could be dealt with in step 4. When a new ARG is generated, it turns into current ARG and a new round begins. For more details, see the Methods section.
Figure 8
A schematic diagram of the update steps of SMC, SMC′, and MaCS under our framework. Regardless of which branch the new branch coalesces into in Step 4, a new ARG is constructed.
Figure 9
Generation of sample-consistent ARG with SC-sample. A) Generation of a binary tree is consistent with the left site. B) Determine whether the current local tree is consistent with the second site, the answer is yes since there is only one mutant edge. C) Determine whether the current local tree is consistent with the third site. This tree is not consistent because there are two mutant edges, so P1 = 0.5. D) Generate the next recombination point that is uniform on [0, P1] and obtain P2 = 0.4. The dotted lines are the branches onto which the new branches are supposed to coalesce. E) The new branch coalesces and the [0, 0.4] part of the ARG is simulated.
Similar articles
- The SMC' is a highly accurate approximation to the ancestral recombination graph.
Wilton PR, Carmi S, Hobolth A. Wilton PR, et al. Genetics. 2015 May;200(1):343-55. doi: 10.1534/genetics.114.173898. Epub 2015 Mar 17. Genetics. 2015. PMID: 25786855 Free PMC article. - The Promise of Inferring the Past Using the Ancestral Recombination Graph.
Brandt DYC, Huber CD, Chiang CWK, Ortega-Del Vecchyo D. Brandt DYC, et al. Genome Biol Evol. 2024 Feb 1;16(2):evae005. doi: 10.1093/gbe/evae005. Genome Biol Evol. 2024. PMID: 38242694 Free PMC article. - Genome-wide inference of ancestral recombination graphs.
Rasmussen MD, Hubisz MJ, Gronau I, Siepel A. Rasmussen MD, et al. PLoS Genet. 2014 May 15;10(5):e1004342. doi: 10.1371/journal.pgen.1004342. eCollection 2014. PLoS Genet. 2014. PMID: 24831947 Free PMC article. - The era of the ARG: An introduction to ancestral recombination graphs and their significance in empirical evolutionary genomics.
Lewanski AL, Grundler MC, Bradburd GS. Lewanski AL, et al. PLoS Genet. 2024 Jan 18;20(1):e1011110. doi: 10.1371/journal.pgen.1011110. eCollection 2024 Jan. PLoS Genet. 2024. PMID: 38236805 Free PMC article. Review. - Unraveling recombination rate evolution using ancestral recombination maps.
Munch K, Schierup MH, Mailund T. Munch K, et al. Bioessays. 2014 Sep;36(9):892-900. doi: 10.1002/bies.201400047. Epub 2014 Jul 14. Bioessays. 2014. PMID: 25043668 Free PMC article. Review.
Cited by
- Bayesian inference of ancestral recombination graphs.
Mahmoudi A, Koskela J, Kelleher J, Chan YB, Balding D. Mahmoudi A, et al. PLoS Comput Biol. 2022 Mar 9;18(3):e1009960. doi: 10.1371/journal.pcbi.1009960. eCollection 2022 Mar. PLoS Comput Biol. 2022. PMID: 35263345 Free PMC article. - Efficient ancestry and mutation simulation with msprime 1.0.
Baumdicker F, Bisschop G, Goldstein D, Gower G, Ragsdale AP, Tsambos G, Zhu S, Eldon B, Ellerman EC, Galloway JG, Gladstein AL, Gorjanc G, Guo B, Jeffery B, Kretzschumar WW, Lohse K, Matschiner M, Nelson D, Pope NS, Quinto-Cortés CD, Rodrigues MF, Saunack K, Sellinger T, Thornton K, van Kemenade H, Wohns AW, Wong Y, Gravel S, Kern AD, Koskela J, Ralph PL, Kelleher J. Baumdicker F, et al. Genetics. 2022 Mar 3;220(3):iyab229. doi: 10.1093/genetics/iyab229. Genetics. 2022. PMID: 34897427 Free PMC article. - The Multispecies Coalescent Model Outperforms Concatenation Across Diverse Phylogenomic Data Sets.
Jiang X, Edwards SV, Liu L. Jiang X, et al. Syst Biol. 2020 Jul 1;69(4):795-812. doi: 10.1093/sysbio/syaa008. Syst Biol. 2020. PMID: 32011711 Free PMC article. - The Bacterial Sequential Markov Coalescent.
De Maio N, Wilson DJ. De Maio N, et al. Genetics. 2017 May;206(1):333-343. doi: 10.1534/genetics.116.198796. Epub 2017 Mar 3. Genetics. 2017. PMID: 28258183 Free PMC article. - FREQUENT SUBGRAPH MINING OF PERSONALIZED SIGNALING PATHWAY NETWORKS GROUPS PATIENTS WITH FREQUENTLY DYSREGULATED DISEASE PATHWAYS AND PREDICTS PROGNOSIS.
Durmaz A, Henderson TAD, Brubaker D, Bebek G. Durmaz A, et al. Pac Symp Biocomput. 2017;22:402-413. doi: 10.1142/9789813207813_0038. Pac Symp Biocomput. 2017. PMID: 27896993 Free PMC article.
References
- Kingman JFC. On the genealogy of large populations. J Appl Probab. 1982;19A:27–43. doi: 10.2307/3213548. - DOI
- Kingman JFC. The coalescent. Stoch ProcessAppl. 1982;13:235–248. doi: 10.1016/0304-4149(82)90011-4. - DOI
- Griffiths RCaM P. An ancestral recombination graph. Prog Popul Genet Hum Evol. 1997;87:257–270. doi: 10.1007/978-1-4757-2609-1_16. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous