HTSeq--a Python framework to work with high-throughput sequencing data - PubMed (original) (raw)
HTSeq--a Python framework to work with high-throughput sequencing data
Simon Anders et al. Bioinformatics. 2015.
Abstract
Motivation: A large choice of tools exists for many standard tasks in the analysis of high-throughput sequencing (HTS) data. However, once a project deviates from standard workflows, custom scripts are needed.
Results: We present HTSeq, a Python library to facilitate the rapid development of such scripts. HTSeq offers parsers for many common data formats in HTS projects, as well as classes to represent data, such as genomic coordinates, sequences, sequencing reads, alignments, gene model information and variant calls, and provides data structures that allow for querying via genomic coordinates. We also present htseq-count, a tool developed with HTSeq that preprocesses RNA-Seq data for differential expression analysis by counting the overlap of reads with genes.
Availability and implementation: HTSeq is released as an open-source software under the GNU General Public Licence and available from http://www-huber.embl.de/HTSeq or from the Python Package Index at https://pypi.python.org/pypi/HTSeq.
© The Author 2014. Published by Oxford University Press.
Figures
Fig. 1.
(a) The SAM_Alignment class as an example of an HTSeq data record: subsets of the content are bundled in object-valued fields, using classes (here SequenceWithQualities and GenomicInterval) that are also used in other data records to provide a common view on diverse data types. (b) The cigar field in a SAM_alignment object presents the detailed structure of a read alignment as a list of CigarOperation. This allows for convenient downstream processing of complicated alignment structures, such as the one given by the cigar string on top and illustrated in the middle. Five CigarOperation objects, with slots for the columns of the table (bottom) provide the data from the cigar string, along with the inferred coordinates of the affected regions in read (‘query’) and reference
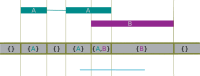
Fig. 2.
Using the class GenomicArrayOfSets to represent overlapping annotation metadata. The indicated features are assigned to the array, which then represents them internally as steps, each step having as value a set whose elements are references to the features overlapping the step
Similar articles
- Analysing high-throughput sequencing data in Python with HTSeq 2.0.
Putri GH, Anders S, Pyl PT, Pimanda JE, Zanini F. Putri GH, et al. Bioinformatics. 2022 May 13;38(10):2943-2945. doi: 10.1093/bioinformatics/btac166. Bioinformatics. 2022. PMID: 35561197 Free PMC article. - htseq-clip: a toolset for the preprocessing of eCLIP/iCLIP datasets.
Sahadevan S, Sekaran T, Ashaf N, Fritz M, Hentze MW, Huber W, Schwarzl T. Sahadevan S, et al. Bioinformatics. 2023 Jan 1;39(1):btac747. doi: 10.1093/bioinformatics/btac747. Bioinformatics. 2023. PMID: 36394253 Free PMC article. - Rcount: simple and flexible RNA-Seq read counting.
Schmid MW, Grossniklaus U. Schmid MW, et al. Bioinformatics. 2015 Feb 1;31(3):436-7. doi: 10.1093/bioinformatics/btu680. Epub 2014 Oct 15. Bioinformatics. 2015. PMID: 25322836 - Omics Pipe: a community-based framework for reproducible multi-omics data analysis.
Fisch KM, Meißner T, Gioia L, Ducom JC, Carland TM, Loguercio S, Su AI. Fisch KM, et al. Bioinformatics. 2015 Jun 1;31(11):1724-8. doi: 10.1093/bioinformatics/btv061. Epub 2015 Jan 30. Bioinformatics. 2015. PMID: 25637560 Free PMC article. - Bioinformatics tools for analysing viral genomic data.
Orton RJ, Gu Q, Hughes J, Maabar M, Modha S, Vattipally SB, Wilkie GS, Davison AJ. Orton RJ, et al. Rev Sci Tech. 2016 Apr;35(1):271-85. doi: 10.20506/rst.35.1.2432. Rev Sci Tech. 2016. PMID: 27217183 Review.
Cited by
- Integrated multi-omics identifies pathways governing interspecies interaction between A. fumigatus and K. pneumoniae.
Bitencourt T, Nogueira F, Jenull S, Phan-Canh T, Tscherner M, Kuchler K, Lion T. Bitencourt T, et al. Commun Biol. 2024 Nov 12;7(1):1496. doi: 10.1038/s42003-024-07145-x. Commun Biol. 2024. PMID: 39533021 Free PMC article. - Novel Ser74 of NF-κB/IκBα phosphorylated by MAPK/ERK regulates temperature adaptation in oysters.
Wang C, Jiang Z, Du M, Cong R, Wang W, Zhang T, Chen J, Zhang G, Li L. Wang C, et al. Cell Commun Signal. 2024 Nov 11;22(1):539. doi: 10.1186/s12964-024-01923-0. Cell Commun Signal. 2024. PMID: 39529137 Free PMC article. - Dosage-sensitive maternal siRNAs determine hybridization success in Capsella.
Dziasek K, Santos-González J, Wang K, Qiu Y, Zhu J, Rigola D, Nijbroek K, Köhler C. Dziasek K, et al. Nat Plants. 2024 Nov 11. doi: 10.1038/s41477-024-01844-3. Online ahead of print. Nat Plants. 2024. PMID: 39528633 - Unveiling the impact of hypodermis on gene expression for advancing bioprinted full-thickness 3D skin models.
Avelino TM, Harb SV, Adamoski D, Oliveira LCM, Horinouchi CDS, Azevedo RJ, Azoubel RA, Thomaz VK, Batista FAH, d'Ávila MA, Granja PL, Figueira ACM. Avelino TM, et al. Commun Biol. 2024 Nov 11;7(1):1437. doi: 10.1038/s42003-024-07106-4. Commun Biol. 2024. PMID: 39528562 Free PMC article. - An Optimized Method for Reconstruction of Transcriptional Regulatory Networks in Bacteria Using ChIP-exo and RNA-seq Datasets.
Jang M, Park JY, Lee G, Kim D. Jang M, et al. J Microbiol. 2024 Nov 11. doi: 10.1007/s12275-024-00181-6. Online ahead of print. J Microbiol. 2024. PMID: 39527186
References
- Beazley DM, et al. Proceedings of the 4th USENIX Tcl/Tk workshop. 1996. SWIG: an easy to use tool for integrating scripting languages with C and C++ pp. 129–139.
- Behnel S, et al. Cython: the best of both worlds. Comput. Sci. Eng. 2011;13:31–39.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources