Nanoscale adhesion forces of Pseudomonas aeruginosa type IV Pili - PubMed (original) (raw)
. 2014 Oct 28;8(10):10723-33.
doi: 10.1021/nn5044383. Epub 2014 Oct 6.
Affiliations
- PMID: 25286300
- PMCID: PMC4212785
- DOI: 10.1021/nn5044383
Nanoscale adhesion forces of Pseudomonas aeruginosa type IV Pili
Audrey Beaussart et al. ACS Nano. 2014.
Abstract
A variety of bacterial pathogens use nanoscale protein fibers called type IV pili to mediate cell adhesion, a primary step leading to infection. Currently, how these nanofibers respond to mechanical stimuli and how this response is used to control adhesion is poorly understood. Here, we use atomic force microscopy techniques to quantify the forces guiding the adhesion of Pseudomonas aeruginosa type IV pili to surfaces. Using chemical force microscopy and single-cell force spectroscopy, we show that pili strongly bind to hydrophobic surfaces in a time-dependent manner, while they weakly bind to hydrophilic surfaces. Individual nanofibers are capable of withstanding forces up to 250 pN, thereby explaining how they can resist mechanical stress. Pulling on individual pili yields constant force plateaus, presumably reflecting conformational changes, as well as nanospring properties that may help bacteria to withstand physiological shear forces. Analysis of mutant strains demonstrates that these mechanical responses originate solely from type IV pili, while flagella and the cell surface localized and proposed pili-associated adhesin PilY1 play no direct role. We also demonstrate that bacterial-host interactions involve constant force plateaus, the extension of bacterial pili, and the formation of membrane tethers from host cells. We postulate that the unique mechanical responses of type IV pili unravelled here enable the bacteria to firmly attach to biotic and abiotic surfaces and thus maintain attachment when subjected to high shear forces under physiological conditions, helping to explain why pili play a critical role in colonization of the host.
Keywords: AFM; Pseudomonas aeruginosa; adhesion; chemical force microscopy; mechanics; pathogens; single-cell force spectroscopy; type IV pili.
Figures
Figure 1
Quantifying the nanomechanics of type IV pili on living bacteria using chemical force microscopy. (a,b) AFM deflection images of wild-type P. aeruginosa bacteria recorded in buffer (a) and in air (b), showing that surface appendages can only be visualized in air. Adhesion force histogram (c) and rupture length histogram (d) obtained by recording force curves in M63 medium between a hydrophobic tip and the polar region of a wild-type P. aeruginosa cell (n = 1024 curves). Adhesion values in (c) correspond to the largest adhesion forces observed in each curve, while rupture lengths in (d) correspond to the last rupture events. All curves were obtained using a contact time of 100 ms, a maximum applied force of 250 pN, and approach and retraction speeds of 1.0 μm s–1. Similar data were obtained using three different tips and three cells from different cultures. (e,f) Stretching individual pili yields constant force plateaus: (e) typical plateau curves composed of a region at zero force followed by a progressive, nonlinear increase in the force (red arrows) to reach the constant force regime, and (f) histogram of the average plateau forces (n = 90 curves from three experiments). As illustrated in the right panel, force plateau signatures are believed to result from force-induced conformational changes within the pili. (g,h) Stretched pili also show single linear force peaks, indicating that they behave as nanosprings: typical linear force peak signatures (g) and histogram of maximum adhesion forces (n = 61 curves from three experiments) (h) as well as quantification of spring-like properties, estimation of pilus spring constant _k_p (h; inset). Superimposition of 10 curves shows that spring-like properties are highly reproducible.
Figure 2
Control experiments demonstrate that constant force plateaus and nanospring behaviors originate from type IV pili. Representative forces curves obtained in M63 medium between hydrophobic tips and the polar region of different mutant strains of P. aeruginosa: (a) Δ_flgK_ strain (7% plateau forces and 12% single adhesions; n = 1024 curves), (b) Δ_pilA flgK_::Tn5 strain (0% adhesion), (c) WT/pPilY1 strain (6% plateau forces and 7% single adhesions), and (d) Δ_pilY1 flgK::_Tn5 strain (0% adhesion). All curves were obtained using a contact time of 100 ms, a maximum applied force of 250 pN, and approach and retraction speeds of 1.0 μm s–1. Similar data were obtained using three different tips and three cells from different cultures.
Figure 3
Single-cell force spectroscopy of the interaction between P. aeruginosa and hydrophobic substrates. (a, Inset) To quantify single-cell adhesion forces, living P. aeruginosa bacteria (blue) were attached on polydopamine-coated colloidal probes (pink). (a,c) Adhesion force histograms, (b,d) rupture length histograms, and representative retraction force profiles obtained by recording multiple force–distance curves between three different WT cells (red, green, and blue) and hydrophobic substrates at 100 ms contact time (a,b; n = 495, 957, and 442 curves for each cell) and 1.1 s contact time (c,d; n = 396, 177, and 863 curves). The inset in (b) shows an enlarged view of the force plateau signatures. The curves were obtained using a maximum applied force of 250 pN and approach and retraction speeds of 1.0 μm s–1.
Figure 4
Role of pili and substrate chemistry in controlling P. aeruginosa single-cell adhesion. (a,c) Adhesion force histograms, (b,d) rupture length histograms, and representative retraction force profiles obtained by recording multiple force–distance curves between cells from the pili-less mutant (Δ_pilY1 flgK_::Tn5) and hydrophobic substrata at 100 ms (a,b; n = 906, 539, and 502 curves for each cell) and 1.1 s (c,d; n = 132, 592, 320 curves) contact time. (e,g) Adhesion force histograms, (f,h) rupture length histograms, and representative retraction force profiles obtained by recording multiple force–distance curves between WT cells and hydrophilic substrata at 100 ms (e,f; n = 159, 250, 217 curves for each cell) and 1.1 s (g,h; n = 211, 179, and 251 curves for each cell) contact time.
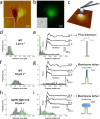
Figure 5
Single-cell force spectroscopy of the P. aeruginosa_–host interaction. (a) AFM deflection image a living A549 pneumocyte cell in buffer (inset: higher magnification of the smooth area). (b) Correlative fluorescence image (inset: corresponding DIC image) of the cell labeled with the fluorescent dye CFSE (green). (c) SCFS was used to quantify the adhesion forces between single P. aeruginosa bacteria (blue) and pneumocytes. For each pneumocyte tested, force curves were recorded on at least three different spots. (d,f) Adhesion force histograms, (e,g) rupture length histograms, and representative retraction force profiles obtained by recording multiple force–distance curves at 1.1 s contact time between WT cells and A549 pneumocytes using a pulling speed of 1.0 μm s–1 (d,e; n = 192 and 434 curves for each cell) and 10.0 μm s–1 (f,g; n = 231 and 223 curves). (h,i) Force data obtained for Δ_pilY1 flgK::Tn5 mutant bacteria at 10.0 μm s–1 pulling speed (n = 180 and 146 curves). As illustrated in the cartoons, long-range constant force plateau interactions of two types are observed with WT bacteria: “rough” plateaus reflecting the extension of bacterial pili (e, right panel) and “smooth” plateaus associated with the extraction of host membrane tethers (g, right panel). With Δ_pilY1 flgK_::Tn5 bacteria, however, only smooth plateaus are observed (i, right panel).
Similar articles
- Nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa pili by using atomic force microscopy.
Touhami A, Jericho MH, Boyd JM, Beveridge TJ. Touhami A, et al. J Bacteriol. 2006 Jan;188(2):370-7. doi: 10.1128/JB.188.2.370-377.2006. J Bacteriol. 2006. PMID: 16385026 Free PMC article. - Single-cell force spectroscopy of pili-mediated adhesion.
Sullan RM, Beaussart A, Tripathi P, Derclaye S, El-Kirat-Chatel S, Li JK, Schneider YJ, Vanderleyden J, Lebeer S, Dufrêne YF. Sullan RM, et al. Nanoscale. 2014 Jan 21;6(2):1134-43. doi: 10.1039/c3nr05462d. Nanoscale. 2014. PMID: 24296882 - Atomic force microscopy analysis of Pel polysaccharide- and type IV pili-mediated adhesion of Pseudomonas aeruginosa PA14 to an abiotic surface.
Beaussart A, Paiva TO, Geiger CJ, Baker AE, O'Toole GA, Dufrêne YF. Beaussart A, et al. Nanoscale. 2024 Jun 27;16(25):12134-12141. doi: 10.1039/d4nr01415d. Nanoscale. 2024. PMID: 38832761 - Assessing bacterial adhesion on an individual adhesin and single pili level using optical tweezers.
Axner O, Andersson M, Björnham O, Castelain M, Klinth J, Koutris E, Schedin S. Axner O, et al. Adv Exp Med Biol. 2011;715:301-13. doi: 10.1007/978-94-007-0940-9_19. Adv Exp Med Biol. 2011. PMID: 21557072 Review. - Bacterial mechanosensing: the force will be with you, always.
Gordon VD, Wang L. Gordon VD, et al. J Cell Sci. 2019 Apr 3;132(7):jcs227694. doi: 10.1242/jcs.227694. J Cell Sci. 2019. PMID: 30944157 Free PMC article. Review.
Cited by
- Adhesion to nanofibers drives cell membrane remodeling through one-dimensional wetting.
Charles-Orszag A, Tsai FC, Bonazzi D, Manriquez V, Sachse M, Mallet A, Salles A, Melican K, Staneva R, Bertin A, Millien C, Goussard S, Lafaye P, Shorte S, Piel M, Krijnse-Locker J, Brochard-Wyart F, Bassereau P, Duménil G. Charles-Orszag A, et al. Nat Commun. 2018 Oct 25;9(1):4450. doi: 10.1038/s41467-018-06948-x. Nat Commun. 2018. PMID: 30361638 Free PMC article. - A double-edged sword: the role of VEGF in wound repair and chemoattraction of opportunist pathogens.
Birkenhauer E, Neethirajan S. Birkenhauer E, et al. Int J Mol Sci. 2015 Mar 30;16(4):7159-72. doi: 10.3390/ijms16047159. Int J Mol Sci. 2015. PMID: 25830483 Free PMC article. - High-Throughput Screen for Inhibitors of the Type IV Pilus Assembly ATPase PilB.
Dye KJ, Vogelaar NJ, Sobrado P, Yang Z. Dye KJ, et al. mSphere. 2021 Mar 3;6(2):e00129-21. doi: 10.1128/mSphere.00129-21. mSphere. 2021. PMID: 33658276 Free PMC article. - Self-organized canals enable long-range directed material transport in bacterial communities.
Li Y, Liu S, Zhang Y, Seng ZJ, Xu H, Yang L, Wu Y. Li Y, et al. Elife. 2022 Sep 26;11:e79780. doi: 10.7554/eLife.79780. Elife. 2022. PMID: 36154945 Free PMC article. - How Microbes Use Force To Control Adhesion.
Viljoen A, Mignolet J, Viela F, Mathelié-Guinlet M, Dufrêne YF. Viljoen A, et al. J Bacteriol. 2020 May 27;202(12):e00125-20. doi: 10.1128/JB.00125-20. Print 2020 May 27. J Bacteriol. 2020. PMID: 32253344 Free PMC article. Review.
References
- Costerton J. W.; Stewart P. S.; Greenberg E. P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. - PubMed
- Douglas L. J. Candida Biofilms and Their Role in Infection. Trends Microbiol. 2003, 11, 30–36. - PubMed
- Kolter R.; Greenberg E. P. Microbial Sciences: The Superficial Life of Microbes. Nature 2006, 441, 300–302. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous