Meta-analyses of human gut microbes associated with obesity and IBD - PubMed (original) (raw)
Meta-Analysis
Meta-analyses of human gut microbes associated with obesity and IBD
William A Walters et al. FEBS Lett. 2014.
Abstract
Recent studies have linked human gut microbes to obesity and inflammatory bowel disease, but consistent signals have been difficult to identify. Here we test for indicator taxa and general features of the microbiota that are generally consistent across studies of obesity and of IBD, focusing on studies involving high-throughput sequencing of the 16S rRNA gene (which we could process using a common computational pipeline). We find that IBD has a consistent signature across studies and allows high classification accuracy of IBD from non-IBD subjects, but that although subjects can be classified as lean or obese within each individual study with statistically significant accuracy, consistent with the ability of the microbiota to experimentally transfer this phenotype, signatures of obesity are not consistent between studies even when the data are analyzed with consistent methods. The results suggest that correlations between microbes and clinical conditions with different effect sizes (e.g. the large effect size of IBD versus the small effect size of obesity) may require different cohort selection and analysis strategies.
Keywords: Gut microbiota; Metaanalysis.
Copyright © 2014 The Authors. Published by Elsevier B.V. All rights reserved.
Figures
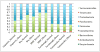
Figure 1. Relative abundance of phylum-level gut microbial taxa
Studies listed below are Zupancic [25], Wu [28], Human microbiome project [44], Turnbaugh [8], and Yatsunenko[45].
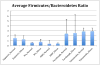
Figure 2
Ratios of Firmicutes:Bacteroidetes in normal versus obese BMI subjects. Means of ratios for each study/BMI category are shown. Error bars are standard error of the mean. The Turnbaugh study includes a number of samples with extremely low Bacteroidetes, leading to large standard error values.
Figure 3
Alpha diversity (observed species) across studies. Metric is observed species (counts of unique OTUs). Sequence depth is 1000 sequences per sample, and subsampling was performed 10 times. P-values were calculated by using a Monte Carlo simulation with 999 permutations
Figure 3
Alpha diversity (observed species) across studies. Metric is observed species (counts of unique OTUs). Sequence depth is 1000 sequences per sample, and subsampling was performed 10 times. P-values were calculated by using a Monte Carlo simulation with 999 permutations
Figure 3
Alpha diversity (observed species) across studies. Metric is observed species (counts of unique OTUs). Sequence depth is 1000 sequences per sample, and subsampling was performed 10 times. P-values were calculated by using a Monte Carlo simulation with 999 permutations
Figure 4
Alpha diversity (Shannon) across studies. Metric is Shannon (abundance and evenness). Sequence depth is 1000 sequences per sample, and subsampling was performed 10 times. P-values were calculated by using a Monte Carlo simulation with 999 permutations
Figure 4
Alpha diversity (Shannon) across studies. Metric is Shannon (abundance and evenness). Sequence depth is 1000 sequences per sample, and subsampling was performed 10 times. P-values were calculated by using a Monte Carlo simulation with 999 permutations
Figure 4
Alpha diversity (Shannon) across studies. Metric is Shannon (abundance and evenness). Sequence depth is 1000 sequences per sample, and subsampling was performed 10 times. P-values were calculated by using a Monte Carlo simulation with 999 permutations
Figure 4
Alpha diversity (Shannon) across studies. Metric is Shannon (abundance and evenness). Sequence depth is 1000 sequences per sample, and subsampling was performed 10 times. P-values were calculated by using a Monte Carlo simulation with 999 permutations
Figure 5
Alpha diversity (PD) for Turnbaugh et al data [8] across clustering methods. Metric is phylogenetic diversity (a measure of branch length of the phylogenetic tree occupied by the sequences present in the samples). Sequence depth is 1000 sequences per sample, and subsampling was performed 10 times. P-values were calculated by using a Monte Carlo simulation with 999 permutations
Figure 5
Alpha diversity (PD) for Turnbaugh et al data [8] across clustering methods. Metric is phylogenetic diversity (a measure of branch length of the phylogenetic tree occupied by the sequences present in the samples). Sequence depth is 1000 sequences per sample, and subsampling was performed 10 times. P-values were calculated by using a Monte Carlo simulation with 999 permutations
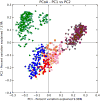
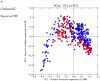
Figure 6. Clustering of BMI samples with unweighted UniFrac
Shape/Color: Study/BMI category Purple square: Zupancic normal Brown triangle: Zupancic obese Orange square: Turnbaugh normal Pink triangle: Turnbaugh obese Red circle: Wu normal Yellow diamond: Wu obese Dark blue circle: HMP normal Light blue triangle: HMP obese Green triangle: Yatsunenko normal Grey circle: Yatsunenko obese
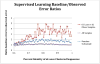
Figure 7. Comparison of supervised learning error ratios to clustering identity of data
Samples matching those used in Knights et al [9] replicated the improved classifications relative to random guessing (value of 1) for lean and obese subjects in the Turnbaugh [8] study, and are shown as the red line. The average error ratio for a random subsample of 30 obese and lean samples (10× sample at each percent identity, average ratio is shown) is depicted in blue. The purple line shows the classification error ratio when all samples (61 lean versus 196 obese samples), which is essentially no better than random guess for any clustering identity. The sequences were clustered using a de novo approach for each percent identity listed.
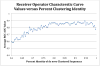
Figure 8. Receiver operator characteristic curve values for all Turnbaugh lean and obese subjects
The average ROC area under the curve value (using random forest method) for each clustering identity was calculated by the averaging the 5× repeated (with 10-fold cross-validation) optimized ROC values. A 0.5 value indicates no better than random guess, while 1.0 indicates perfect sensitivity and specificity. The sequences were clustered using a de novo approach for each percent identity listed.
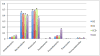
Figure 9. Phylum-level taxa plots for IBD subjects versus healthy controls
Seven most abundant phyla shown. HC=healthy controls, UC=ulcerative colitis, CCD=colonic Crohn's disease, ICD=ileal Crohn's disease. Error bars indicate standard error of the mean.
Figure 10. PCoA plots of healthy controls versus subjects with IBD
Distances were calculated with unweighted UniFrac. A-HC vs UC samples B. HC vs CCD samples C. HC vs ICD samples. Distances between healthy controls and all IBD categories are significantly different (p-value < 0.050 with PERMANOVA tests (999 permutations). Data were evenly sampled at 1004 sequences per sample.
Figure 10. PCoA plots of healthy controls versus subjects with IBD
Distances were calculated with unweighted UniFrac. A-HC vs UC samples B. HC vs CCD samples C. HC vs ICD samples. Distances between healthy controls and all IBD categories are significantly different (p-value < 0.050 with PERMANOVA tests (999 permutations). Data were evenly sampled at 1004 sequences per sample.
Figure 10. PCoA plots of healthy controls versus subjects with IBD
Distances were calculated with unweighted UniFrac. A-HC vs UC samples B. HC vs CCD samples C. HC vs ICD samples. Distances between healthy controls and all IBD categories are significantly different (p-value < 0.050 with PERMANOVA tests (999 permutations). Data were evenly sampled at 1004 sequences per sample.
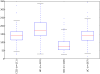
Figure 11. Alpha diversity for IBD subjects and healthy controls
Y-axis indicates observed species value. Healthy control samples are significantly different from all inflammatory bowel disease categories with a p-value of < 0.05 (Monte Carlo permutation test, permutations = 999). The samples were repeatedly sampled (10×) at 1000 sequences/sample.
Similar articles
- Fungal Signature in the Gut Microbiota of Pediatric Patients With Inflammatory Bowel Disease.
Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, Baldassano RN, Lewis JD, Bushman FD, Wu GD. Chehoud C, et al. Inflamm Bowel Dis. 2015 Aug;21(8):1948-56. doi: 10.1097/MIB.0000000000000454. Inflamm Bowel Dis. 2015. PMID: 26083617 Free PMC article. - Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease.
Greenblum S, Turnbaugh PJ, Borenstein E. Greenblum S, et al. Proc Natl Acad Sci U S A. 2012 Jan 10;109(2):594-9. doi: 10.1073/pnas.1116053109. Epub 2011 Dec 19. Proc Natl Acad Sci U S A. 2012. PMID: 22184244 Free PMC article. - The Gut Microbiota in Collagenous Colitis Shares Characteristics With Inflammatory Bowel Disease-Associated Dysbiosis.
Carstens A, Dicksved J, Nelson R, Lindqvist M, Andreasson A, Bohr J, Tysk C, Talley NJ, Agréus L, Engstrand L, Halfvarson J. Carstens A, et al. Clin Transl Gastroenterol. 2019 Jul;10(7):e00065. doi: 10.14309/ctg.0000000000000065. Clin Transl Gastroenterol. 2019. PMID: 31343467 Free PMC article. - The Human Gut Microbiota.
Harmsen HJ, de Goffau MC. Harmsen HJ, et al. Adv Exp Med Biol. 2016;902:95-108. doi: 10.1007/978-3-319-31248-4_7. Adv Exp Med Biol. 2016. PMID: 27161353 Review. - [Fecal microbiota transplantation in treatment of inflammatory bowel diseases].
Privalov MA, Sizenko AK. Privalov MA, et al. Lik Sprava. 2014 Nov;(11):15-21. Lik Sprava. 2014. PMID: 25528828 Review. Russian.
Cited by
- Identification of microbial markers across populations in early detection of colorectal cancer.
Wu Y, Jiao N, Zhu R, Zhang Y, Wu D, Wang AJ, Fang S, Tao L, Li Y, Cheng S, He X, Lan P, Tian C, Liu NN, Zhu L. Wu Y, et al. Nat Commun. 2021 May 24;12(1):3063. doi: 10.1038/s41467-021-23265-y. Nat Commun. 2021. PMID: 34031391 Free PMC article. - Environmental Influences on the Human Microbiome and Implications for Noncommunicable Disease.
Ahn J, Hayes RB. Ahn J, et al. Annu Rev Public Health. 2021 Apr 1;42:277-292. doi: 10.1146/annurev-publhealth-012420-105020. Annu Rev Public Health. 2021. PMID: 33798404 Free PMC article. Review. - When a Neonate Is Born, So Is a Microbiota.
Coscia A, Bardanzellu F, Caboni E, Fanos V, Peroni DG. Coscia A, et al. Life (Basel). 2021 Feb 16;11(2):148. doi: 10.3390/life11020148. Life (Basel). 2021. PMID: 33669262 Free PMC article. Review. - Effect of postnatal low-dose exposure to environmental chemicals on the gut microbiome in a rodent model.
Hu J, Raikhel V, Gopalakrishnan K, Fernandez-Hernandez H, Lambertini L, Manservisi F, Falcioni L, Bua L, Belpoggi F, L Teitelbaum S, Chen J. Hu J, et al. Microbiome. 2016 Jun 14;4(1):26. doi: 10.1186/s40168-016-0173-2. Microbiome. 2016. PMID: 27301250 Free PMC article. - Gut Microbiota in the First 2 Years of Life and the Association with Body Mass Index at Age 12 in a Norwegian Birth Cohort.
Stanislawski MA, Dabelea D, Wagner BD, Iszatt N, Dahl C, Sontag MK, Knight R, Lozupone CA, Eggesbø M. Stanislawski MA, et al. mBio. 2018 Oct 23;9(5):e01751-18. doi: 10.1128/mBio.01751-18. mBio. 2018. PMID: 30352933 Free PMC article.
References
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. - PubMed
Publication types
MeSH terms
Grants and funding
- P01 DK078669/DK/NIDDK NIH HHS/United States
- R01 MD011389/MD/NIMHD NIH HHS/United States
- T32 GM008759/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 HG004872/HG/NHGRI NIH HHS/United States
- T32 GM142607/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical