Structural identification of the Vps18 β-propeller reveals a critical role in the HOPS complex stability and function - PubMed (original) (raw)
Structural identification of the Vps18 β-propeller reveals a critical role in the HOPS complex stability and function
Heide Behrmann et al. J Biol Chem. 2014.
Abstract
Membrane fusion at the vacuole, the lysosome equivalent in yeast, requires the HOPS tethering complex, which is recruited by the Rab7 GTPase Ypt7. HOPS provides a template for the assembly of SNAREs and thus likely confers fusion at a distinct position on vacuoles. Five of the six subunits in HOPS have a similar domain prediction with strong similarity to COPII subunits and nuclear porins. Here, we show that Vps18 indeed has a seven-bladed β-propeller as its N-terminal domain by revealing its structure at 2.14 Å. The Vps18 N-terminal domain can interact with the N-terminal part of Vps11 and also binds to lipids. Although deletion of the Vps18 N-terminal domain does not preclude HOPS assembly, as revealed by negative stain electron microscopy, the complex is instable and cannot support membrane fusion in vitro. We thus conclude that the β-propeller of Vps18 is required for HOPS stability and function and that it can serve as a starting point for further structural analyses of the HOPS tethering complex.
Keywords: Crystal Structure; Endosome; HOPS; Membrane Fusion; Rab; SNARE Proteins; Tethering Complex; Vacuole.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
FIGURE 1.
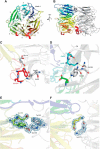
Vps18NTD folds into a seven-bladed β-propeller. A schematic representation front view (A) and side view (B) of the crystal dimer is shown. The propeller blades of chain B are colored individually, whereas chain A is depicted in gray. Details are of the crystal dimer interface. Shown are stick representations of regions highlighted in Fig. 4. C and D corresponds to box D1 and D1′ of Fig. 4, E to D2, and F to D3. The blue mesh depicts the 2m|Fo| − D|Fc| electron density contoured at 1 σ. Yellow dashed lines indicate hydrogen bonds.
FIGURE 2.
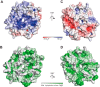
The interface of the crystal dimer is dominated by polar interactions. A–D, surface representations of chain B colored by (A and C) electrostatic potential or (B and D) hydrophobic potential.
FIGURE 3.
The Vps18 β-propeller interacts with the predicted Vps11 propeller. A and B, interaction of Vps18NTD-GST with subcomplexes of HOPS. Vps18NTD-GST was incubated with the indicated Vps11-Vps39 dimer, the Vps11-Vps39-Vps18 trimer, with the Vps16-Vps33 dimer, and with the entire HOPS complex carrying a GFP tag on Vps33. The GST pulldown was conducted as before (14). Eluted proteins were analyzed on Coomassie-stained gels (CBB) or by Western blot (WB) against the calmodulin-binding protein (CbP) tag, which is part of the TAP tag. C, purified His-tagged Vps11 β-propeller was incubated with either GST or Vps18NTD-GST. Proteins were eluted by boiling and analyzed on Coomassie-stained gels or Western blot against the His-tag. D and E, Vps11NTD and Vps18NTD bound to membranes. Both domains were incubated with liposomes of different size and then pelleted by centrifugation (30 min at 100,000 g, 4 °C). S, supernatant (gray bars); P, pellet (dark gray bars). F, analysis of charge mutants in Vps18NTD was conducted as in F. AIA corresponds to K97A/K99A, and TIT corresponds to T97A/T99A.
FIGURE 4.
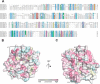
The dimer interface is less conserved than the residues involved in lipid binding. A, sequence alignment of the N-terminal domain of Vps18 across different eukaryotic species. Residues are colored according to the ClustalX scheme, which color codes conserved residues by their chemical nature: hydrophobic in blue, basic in red, polar uncharged in green, acidic in magenta, hydrophilic aromatic in cyan, cysteine in pink, glycine in orange, and proline in yellow. B and C, surface representation of chain B colored by evolutionary conservation. Key residues involved in lipid binding (B) and the crystal dimer interface (C) are highlighted and shown in detail in Fig. 1. Boxes D1 and D1′ correspond to panels C and D of Fig. 1, D2 to E, and D3 to F.
FIGURE 5.
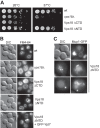
Role of Vps18NTD for endosomal function in yeast cells. A, growth assay of Vps18 mutants. The indicated strains carrying either a deletion of Vps18, wild-type Vps18, or C-terminal and N-terminal truncations were spotted onto yeast extract/peptone/dextrose plates and grown at 26 °C or 37 °C for up to 4 days. B, effect of mutations on vacuole morphology. Vacuoles in the respective strains were stained with 30 μ
m
FM4-64 according to standard protocols (42), and cells were observed by fluorescence microscopy. DIC, differential interference contrast. C, cells lacking the Vps18NTD with endocytic defects. Mup1 was GFP-tagged and monitored upon methionine addition by fluorescence microscopy.
FIGURE 6.
Vps18NTD is required for HOPS stability. A, purification of HOPS without Vps18NTD. HOPS was purified on IgG beads via Vps41 (left) or Vps18 (right) from strains overexpressing all six subunits as described (12, 14). Eluates were applied to SDS-PAGE and Coomassie-stained. B, sizing of HOPS without Vps18NTD. HOPS-Vps18-TAP eluates of A were loaded onto 10–30% glycerol gradients and centrifuged for 18 h at 30,000 rpm in a SW40 rotor at 4 °C. 1-ml fractions were TCA-precipitated and analyzed by SDS-PAGE and Coomassie staining. C, representative electron micrograph of HOPS without Vps18NTD with examples for the different negatively stained particle types boxed. Scale bar, 100 nm. D, diverse particle sizes observed correspond to the dimensions of the complete HOPS without Vps18NTD (1), Vps18ΔNTD-Vps11-Vps39 (2), Vps41 (3), or Vps41- Vps18ΔNTD-Vps11-Vps39 (4). E, the datasets of HOPS-Vps39GFP and the complete HOPS without Vps18NTD were combined, aligned, and classified as described for HOPS-Vps39-GFP (12). Three representative class averages are shown in the third column (each of them containing about 110 particles). Averages calculated from the respective subclasses of HOPS-Vps39-GFP as well as HOPS without Vps18NTD are presented in the first and second columns, respectively. To analyze differences between both subclasses the variance (column 4) was calculated. F, liposome fusion requires the Vps18NTD within HOPS. Proteoliposomes bearing all four SNAREs (Vam3, Vam7, Vti1, and Nyv1) were prepared as described (37). One population carried both NBD (12-(_N_-methyl-_N_-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)))-phosphatidylethanolamine and rhodamine-phosphatidylethanolamine. Fusion was initiated by the addition of Sec17, Sec18, and HOPS (50 n
m
) and monitored by fluorescence dequenching (37).
Similar articles
- HOPS drives vacuole fusion by binding the vacuolar SNARE complex and the Vam7 PX domain via two distinct sites.
Krämer L, Ungermann C. Krämer L, et al. Mol Biol Cell. 2011 Jul 15;22(14):2601-11. doi: 10.1091/mbc.E11-02-0104. Epub 2011 May 25. Mol Biol Cell. 2011. PMID: 21613544 Free PMC article. - The Habc domain of the SNARE Vam3 interacts with the HOPS tethering complex to facilitate vacuole fusion.
Lürick A, Kuhlee A, Bröcker C, Kümmel D, Raunser S, Ungermann C. Lürick A, et al. J Biol Chem. 2015 Feb 27;290(9):5405-13. doi: 10.1074/jbc.M114.631465. Epub 2015 Jan 6. J Biol Chem. 2015. PMID: 25564619 Free PMC article. - Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis.
Takemoto K, Ebine K, Askani JC, Krüger F, Gonzalez ZA, Ito E, Goh T, Schumacher K, Nakano A, Ueda T. Takemoto K, et al. Proc Natl Acad Sci U S A. 2018 Mar 6;115(10):E2457-E2466. doi: 10.1073/pnas.1717839115. Epub 2018 Feb 20. Proc Natl Acad Sci U S A. 2018. PMID: 29463724 Free PMC article. - CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion.
Balderhaar HJ, Ungermann C. Balderhaar HJ, et al. J Cell Sci. 2013 Mar 15;126(Pt 6):1307-16. doi: 10.1242/jcs.107805. J Cell Sci. 2013. PMID: 23645161 Review. - Functional homologies in vesicle tethering.
Kuhlee A, Raunser S, Ungermann C. Kuhlee A, et al. FEBS Lett. 2015 Sep 14;589(19 Pt A):2487-97. doi: 10.1016/j.febslet.2015.06.001. Epub 2015 Jun 10. FEBS Lett. 2015. PMID: 26072291 Review.
Cited by
- Mucopolysaccharidosis-Plus Syndrome.
Vasilev F, Sukhomyasova A, Otomo T. Vasilev F, et al. Int J Mol Sci. 2020 Jan 9;21(2):421. doi: 10.3390/ijms21020421. Int J Mol Sci. 2020. PMID: 31936524 Free PMC article. - A novel lysosome-related prognostic signature associated with prognosis and immune infiltration landscape in oral squamous cell carcinoma.
Liu JJ, Xu ZM, Liu Y, Guo XY, Zhang WB. Liu JJ, et al. Heliyon. 2024 Feb 14;10(4):e26100. doi: 10.1016/j.heliyon.2024.e26100. eCollection 2024 Feb 29. Heliyon. 2024. PMID: 38420448 Free PMC article. - Coat/Tether Interactions-Exception or Rule?
Schroeter S, Beckmann S, Schmitt HD. Schroeter S, et al. Front Cell Dev Biol. 2016 May 17;4:44. doi: 10.3389/fcell.2016.00044. eCollection 2016. Front Cell Dev Biol. 2016. PMID: 27243008 Free PMC article. Review. - Multiple Roles of the Small GTPase Rab7.
Guerra F, Bucci C. Guerra F, et al. Cells. 2016 Aug 18;5(3):34. doi: 10.3390/cells5030034. Cells. 2016. PMID: 27548222 Free PMC article. Review. - Vps11 and Vps18 of Vps-C membrane traffic complexes are E3 ubiquitin ligases and fine-tune signalling.
Segala G, Bennesch MA, Ghahhari NM, Pandey DP, Echeverria PC, Karch F, Maeda RK, Picard D. Segala G, et al. Nat Commun. 2019 Apr 23;10(1):1833. doi: 10.1038/s41467-019-09800-y. Nat Commun. 2019. PMID: 31015428 Free PMC article.
References
- Kümmel D., Ungermann C. (2014) Principles of membrane tethering and fusion in endosome and lysosome biogenesis. Curr. Opin. Cell Biol. 29, 61–66 - PubMed
- Yu I.-M., Hughson F. M. (2010) Tethering factors as organizers of intracellular vesicular traffic. Annu. Rev. Cell Dev. Biol. 26, 137–156 - PubMed
- Zhang X., He X., Fu X.-Y., Chang Z. (2006) Varp is a Rab21 guanine nucleotide exchange factor and regulates endosome dynamics. J. Cell Sci. 119, 1053–1062 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases