Identification of a novel drug lead that inhibits HCV infection and cell-to-cell transmission by targeting the HCV E2 glycoprotein - PubMed (original) (raw)
. 2014 Oct 30;9(10):e111333.
doi: 10.1371/journal.pone.0111333. eCollection 2014.
Laurence Cocquerel 2, Adam Zemla 3, Laure Saas 2, Jean Dubuisson 2, Jost Vielmetter 4, Joseph Marcotrigiano 5, Abdul Ghafoor Khan 5, Felipe Vences Catalan 6, Alexander L Perryman 7, Joel S Freundlich 8, Stefano Forli 9, Shoshana Levy 6, Rod Balhorn 10, Hassan M Azzazy 1
Affiliations
- PMID: 25357246
- PMCID: PMC4214736
- DOI: 10.1371/journal.pone.0111333
Identification of a novel drug lead that inhibits HCV infection and cell-to-cell transmission by targeting the HCV E2 glycoprotein
Reem R Al Olaby et al. PLoS One. 2014.
Abstract
Hepatitis C Virus (HCV) infects 200 million individuals worldwide. Although several FDA approved drugs targeting the HCV serine protease and polymerase have shown promising results, there is a need for better drugs that are effective in treating a broader range of HCV genotypes and subtypes without being used in combination with interferon and/or ribavirin. Recently, two crystal structures of the core of the HCV E2 protein (E2c) have been determined, providing structural information that can now be used to target the E2 protein and develop drugs that disrupt the early stages of HCV infection by blocking E2's interaction with different host factors. Using the E2c structure as a template, we have created a structural model of the E2 protein core (residues 421-645) that contains the three amino acid segments that are not present in either structure. Computational docking of a diverse library of 1,715 small molecules to this model led to the identification of a set of 34 ligands predicted to bind near conserved amino acid residues involved in the HCV E2: CD81 interaction. Surface plasmon resonance detection was used to screen the ligand set for binding to recombinant E2 protein, and the best binders were subsequently tested to identify compounds that inhibit the infection of Huh-7 cells by HCV. One compound, 281816, blocked E2 binding to CD81 and inhibited HCV infection in a genotype-independent manner with IC50's ranging from 2.2 µM to 4.6 µM. 281816 blocked the early and late steps of cell-free HCV entry and also abrogated the cell-to-cell transmission of HCV. Collectively the results obtained with this new structural model of E2c suggest the development of small molecule inhibitors such as 281816 that target E2 and disrupt its interaction with CD81 may provide a new paradigm for HCV treatment.
Conflict of interest statement
Competing Interests: Reem Al Olaby, Dr. Hassan Azzazy and Dr. Rod Balhorn are co-inventors on a patent application related to the described work that has been submitted by The American University in Cairo, Cairo, Egypt. The title for the application is Ligands That Target Hepatitis C Virus E2 Protein. None of the other coauthors have competing interests. This does not alter the authors' adherence to PLOS ONE policies on sharing data and materials.
Figures
Figure 1. Comparison of structural templates used for modeling the HCV E2c protein.
(A) Bar representation of E2 sequence showing the structural similarities between crystal structures 4MWF chains C and D (E2c structure, genotype 1a), and 4NX3 chain D (genotype 2a). Regions reported in the coordinates span amino acid residues from H421 to N645. The percent sequence identities between amino acid sequences taken from coordinates and corresponding sequence fragments from HCV E2 protein of genotype 1a are shown in the column Seq_ID. In green are colored regions where structural deviations are below 3 Ångstroms measured as Cα-Cα distances between corresponding residues from the superimposed structures. In red are regions where structural data is missing or deviations are greater than 3 Angstroms. The locations of amino acid residues that have been reported to be important for E2 binding to CD81 are marked with yellow stars. (B) Structural superposition of 4mwf_C and 4nx3_D shows strong conformational similarities between experimentally solved structures of E2 proteins for which the level of sequence identity is 69%. In blue and purple are colored structural fragments where two structures 4mwf_C (566–601; Blue: light-dark) and 4nx3_D (568–605; Purple: light-dark) significantly differ. (C) Surface presentation of the 4mwf_D structure showing the amino acid residues identified to be important for E2 binding to CD81 (yellow). The other amino acid residues are color coded with the most hydrophilic residues being colored blue, the most hydrophobic residues colored red orange, and intermediate residues colored white.
Figure 2. Comparison of the crystal structure of E2c with the homology model.
Structural superposition between E2c crystal structure from the PDB chain 4mwf_D (red) and the homology model (black) is illustrated using a ribbon representation. The crystal structure and homology model overlap in most of the regions, except the fragments where coordinates in the experimental structure are missing (red dashed lines).
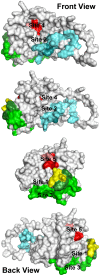
Figure 3. Location of ligand-binding sites on the E2 homology model used to select ligands for testing.
Each of these sites either covers or is located immediately adjacent to amino acids or peptide segments of the E2 protein known to be important for HCV infectivity. H421–N423 (yellow): each amino acid in this region is important for infectivity. Antibodies binding to amino acids Y474–R492 (light cyan) have been shown to prevent infectivity, but this region of the protein has no effect on E2 binding CD81. W487 (dark cyan) is a key amino acid that is involved in E2 binding to E1. S522–G551 (light green) and Y527 and W529 (dark green) are critical for E2 binding to CD81. Site 4: P612, Y613, and H617–P619 (red) are critical for E2 binding to CD81; mutations to R614–W616 (pink) disrupt the structure of the region. The four views show the structure as it is rotated counterclockwise from left to right. Movie S1 shows the rotating structure.
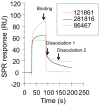
Figure 4. Surface plasmon resonance sensorgrams of ligands binding to recombinant E2 protein (Biacore T100).
This figure shows sensorgrams (binding and dissociation plots) for three of the ligands that bound to the recombinant E2 protein immobilized on a CM5 chip, 281816 (black), 86467 (green) and 121861 (red), and the three reference points that are used to measure the binding and dissociation (dissociation 1 and dissociation 2) of the compound expressed in response units (RU).
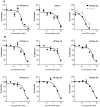
Figure 5. 281816 inhibits HCV entry in a genotype-independent manner.
(A) Huh-7 cells in 96-well plates were pre-treated with 281816 (left and middle panels) or anti-CD81 antibody (right panel) at the indicated concentrations and then infected with HCVpp 2a or HCVcc. (B) Huh-7 cells in 24-well plates were pre-treated with 281816 at the indicated concentrations and infected with HCVpp expressing envelope proteins from the indicated genotype. After 30 hr of infection, cells were lysed and luciferase activities quantified. HCVpp infections were normalized to RD114pp infections.
Figure 6. 281816 inhibits HCV entry.
(A) Huh-7 cells in 24-well plates were treated at different time points with 281816 at 10 µM and infected with HCVcc for 2 hr at 37°C. 281816 was added full-time during the experiment (a), 2 hr before virus inoculation (b), 2 hr during virus inoculation (c), or full-time after virus inoculation (d). (B) Huh-7 cells were infected with HCVcc for 1 hr at 4°C (Step 1: attachment/binding), then virus was removed and cells incubated again at 4°C for 1 hr (Step 2: post-attachment/binding). Finally, cells were shifted at 37°C for 1 hr (Step 3: endocytosis/fusion) and left at 37°C for 21 hr. 281816 was added at 10 µM either during the Step 1, Step 2, Step 3 or Steps 1-2-3. * and *** indicate p values below 0.05 and 0.0001, respectively.
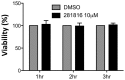
Figure 7. Viability of Huh-7 cells treated with 281816 in the HCV entry experiments.
An MTS assay was used to determine the viability of cells treated with 10 µM 281816 in DMSO (and DMSO alone, as a control) for 1 hr, 2 hr, or 3 hr under the same conditions used in the HCV entry experiments. 281816 is not toxic under any of the conditions used in this assay. There were no significant differences between the 281816 treated and control samples (p values <0.05).
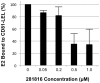
Figure 8. 281816 inhibition of HCV E2 protein binding to native CD81 on Raji cells.
Flow cytometry was used to quantify recombinant HCV E2 protein binding to native CD81 over-expressed on Raji cells. Binding of the recombinant E2 protein to native CD81 on the surface of Raji cells was detected using the mouse monoclonal E2 antibody clone H53 followed by staining with a secondary FITC anti-mouse antibody. E2 binding is inhibited by 281816 in a dose-dependent manner up to 100 µM.
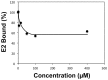
Figure 9. Ligand 281816 inhibits HCV-E2 binding to recombinant CD81-LEL.
Binding of recombinant E2 protein to GST-tagged human CD81-LEL immobilized on a 96 well plate was determined using an ELISA assay. The plate was coated with GST-tagged human CD81-LEL (5 µg/ml) overnight as previously described , HCV E2 protein (5 µg/ml) was pre-incubated with different concentrations of 281816 for 30 min before adding to the plate, and the HCV-E2 protein (with or without the ligand) was then added to the GST-tagged human CD81-LEL coated plate and incubated for 1 hr. HCV-E2 binding was detected using a primary mouse anti-E2 antibody (H53 clone) and a secondary goat anti-mouse-HRP antibody by measuring the absorbance at 405 nm. The results, which are plotted as percent of E2 protein bound to CD81-LEL relative to E2 binding observed in the absence of the ligand (buffer control), show a dose-dependent effect of 281816 on the inhibition of the E2 protein binding to immobilized recombinant CD81-LEL. P values for the 0.05, 0.2, 0.5 and 1.0 µM 281816 samples are 0.0069, 0.0195, 0.0006 and 0.0009 respectively.
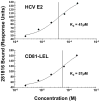
Figure 10. Single cycle kinetics of 281816 binding to recombinant HCV E2 and his-tagged CD81-LEL.
Using surface plasmon resonance detection, ligand 281816 is observed to bind to both HCV E2 and CD81-LEL proteins immobilized on a CM5 chip. Analyses of the binding kinetics were used to obtain dissociation constants for 281816 binding to recombinant E2 (Kd = 41 µM) and recombinant his-tagged CD81-LEL (Kd = 57 µM) protein.
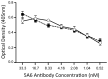
Figure 11. Ligand 281816 does not compete with binding of an anti-CD81 antibody to human CD81-LEL.
Binding of anti-CD81 monoclonal antibody 5A6 to recombinant GST-tagged human CD81-LEL protein was determined by ELISA. Serial dilutions of 5A6 antibody were incubated with the CD81-LEL protein in the presence of 1 µM ligand 281816 (black squares) or PBS as a control (open circles). The amount of 5A6 antibody bound to CD81 remained the same in the presence and absence of the ligand, demonstrating that 281816 does not bind sufficiently well to the E2 binding site on CD81 to block 5A6 binding, even when the antibody concentration was reduced to 1/2000th the concentration of the ligand.
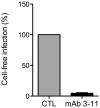
Figure 12. Cell-free infection of Huh-7 cells by HCVcc is blocked by the anti-E2 antibody 3/11.
HCVcc were pre-incubated for 1 hr with neutralizing anti-E2 monoclonal antibody 3/11 at a concentration of 50 µg/ml and next inoculated to Huh-7 cells for 2 hr. Three days post-infection, cells were fixed, stained with the mouse anti-E1 antibody A4 and Alexa555 conjugated anti-mouse IgG, and the number of infected cells was counted. The results show the anti-E2 antibody 3/11 effectively blocks cell-free infection of Huh-7 cells by HCVcc.
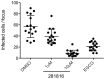
Figure 13. 281816 blocks HCV cell-to-cell transmission.
Huh-7 cells were seeded on coverslips and infected with HCVcc for 2 hr at 37°C. Cells were then washed and cultured for 72 hr at 37°C in culture medium containing the 3/11 neutralizing antibody (50 µg/ml) in presence or in absence of 281816 at indicated concentrations. Cells cultured in presence of DMSO or EGCG at 50 µM were used as controls. The number of infected cells per focus was determined by A4 indirect immunofluorescence. The results show treatment with 281816 significantly reduces the number of infected cells per focus in a dose-dependent manner. Mean p values were below 0.001 for 1 µM 281816 and below 0.0001 for the 10 µM 281816 and EGCG treatment groups.
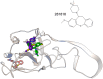
Figure 14. Relative location of 281816 binding sites 1 and 4 on HCV E2.
281816 (structure, top) is predicted to bind to two sites on the E2 protein. Two conformers of 281816 with the lowest free energy of binding are shown bound to site 4. The conformer with the lowest free energy of binding to site 1 is also shown. A video showing the surface structure of the E2 homology model with the three 281816 conformers bound that rotates 360° can be found in Movie S1.
Similar articles
- Identification of ligands that target the HCV-E2 binding site on CD81.
Olaby RA, Azzazy HM, Harris R, Chromy B, Vielmetter J, Balhorn R. Olaby RA, et al. J Comput Aided Mol Des. 2013 Apr;27(4):337-46. doi: 10.1007/s10822-013-9649-3. Epub 2013 Apr 24. J Comput Aided Mol Des. 2013. PMID: 23612915 - Identification of conserved residues in hepatitis C virus envelope glycoprotein E2 that modulate virus dependence on CD81 and SRB1 entry factors.
Lavie M, Sarrazin S, Montserret R, Descamps V, Baumert TF, Duverlie G, Séron K, Penin F, Dubuisson J. Lavie M, et al. J Virol. 2014 Sep;88(18):10584-97. doi: 10.1128/JVI.01402-14. Epub 2014 Jul 2. J Virol. 2014. PMID: 24990994 Free PMC article. - A novel small molecule inhibitor of hepatitis C virus entry.
Baldick CJ, Wichroski MJ, Pendri A, Walsh AW, Fang J, Mazzucco CE, Pokornowski KA, Rose RE, Eggers BJ, Hsu M, Zhai W, Zhai G, Gerritz SW, Poss MA, Meanwell NA, Cockett MI, Tenney DJ. Baldick CJ, et al. PLoS Pathog. 2010 Sep 2;6(9):e1001086. doi: 10.1371/journal.ppat.1001086. PLoS Pathog. 2010. PMID: 20838466 Free PMC article. - [Research on hepatitis C virus entry inhibitor].
Zeng Wenting, Lu X, Wang J, Jin X, Zhu J. Zeng Wenting, et al. Bing Du Xue Bao. 2015 Jan;31(1):97-105. Bing Du Xue Bao. 2015. PMID: 25997338 Review. Chinese. - Unexpected structural features of the hepatitis C virus envelope protein 2 ectodomain.
Sabahi A, Uprichard SL, Wimley WC, Dash S, Garry RF. Sabahi A, et al. J Virol. 2014 Sep;88(18):10280-8. doi: 10.1128/JVI.00874-14. Epub 2014 Jul 2. J Virol. 2014. PMID: 24991010 Free PMC article. Review.
Cited by
- Molecular Mechanisms Underlying the Cellular Entry and Host Range Restriction of Lujo Virus.
Saito T, Hattori T, Okuya K, Manzoor R, Miyamoto H, Kajihara M, Takada A. Saito T, et al. mBio. 2021 Feb 22;13(1):e0306021. doi: 10.1128/mbio.03060-21. Epub 2022 Feb 15. mBio. 2021. PMID: 35164564 Free PMC article. - Targeting of Tetraspanin CD81 with Monoclonal Antibodies and Small Molecules to Combat Cancers and Viral Diseases.
Bailly C, Thuru X. Bailly C, et al. Cancers (Basel). 2023 Apr 6;15(7):2186. doi: 10.3390/cancers15072186. Cancers (Basel). 2023. PMID: 37046846 Free PMC article. Review. - Identification of Potential HCV Inhibitors Based on the Interaction of Epigallocatechin-3-Gallate with Viral Envelope Proteins.
Shahid F, Noreen, Ali R, Badshah SL, Jamal SB, Ullah R, Bari A, Majid Mahmood H, Sohaib M, Akber Ansari S. Shahid F, et al. Molecules. 2021 Feb 26;26(5):1257. doi: 10.3390/molecules26051257. Molecules. 2021. PMID: 33652639 Free PMC article. - Suramin Inhibits Chikungunya Virus Entry and Transmission.
Ho YJ, Wang YM, Lu JW, Wu TY, Lin LI, Kuo SC, Lin CC. Ho YJ, et al. PLoS One. 2015 Jul 24;10(7):e0133511. doi: 10.1371/journal.pone.0133511. eCollection 2015. PLoS One. 2015. PMID: 26208101 Free PMC article. - Charting a Path to Success in Virtual Screening.
Forli S. Forli S. Molecules. 2015 Oct 15;20(10):18732-58. doi: 10.3390/molecules201018732. Molecules. 2015. PMID: 26501243 Free PMC article. Review.
References
- Marks KM, Jacobson IM (2012) The first wave: HCV NS3 protease inhibitors telaprevir and boceprevir. Antivir Ther 17(6 Pt B): 1119–1131. 10.3851/IMP2424; 10.3851/IMP2424. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical