Comparative genomics of Saccharomyces cerevisiae natural isolates for bioenergy production - PubMed (original) (raw)
Comparative Study
Comparative genomics of Saccharomyces cerevisiae natural isolates for bioenergy production
Dana J Wohlbach et al. Genome Biol Evol. 2014 Sep.
Abstract
Lignocellulosic plant material is a viable source of biomass to produce alternative energy including ethanol and other biofuels. However, several factors—including toxic by products from biomass pretreatment and poor fermentation of xylose and other pentose sugars—currently limit the efficiency of microbial biofuel production. To begin to understand the genetic basis of desirable traits, we characterized three strains of Saccharomyces cerevisiae with robust growth in a pretreated lignocellulosic hydrolysate or tolerance to stress conditions relevant to industrial biofuel production, through genome and transcriptome sequencing analysis. All stress resistant strains were highly mosaic, suggesting that genetic admixture may contribute to novel allele combinations underlying these phenotypes. Strain-specific gene sets not found in the lab strain were functionally linked to the tolerances of particular strains. Furthermore,genes with signatures of evolutionary selection were enriched for functional categories important for stress resistance and included stress-responsive signaling factors. Comparison of the strains’ transcriptomic responses to heat and ethanol treatment—two stresses relevant to industrial bioethanol production—pointed to physiological processes that were related to particular stress resistance profiles. Many of the genotype-by-environment expression responses occurred at targets of transcription factors with signatures of positive selection, suggesting that these strains have undergone positive selection for stress tolerance. Our results generate new insights into potential mechanisms of tolerance to stresses relevant to biofuel production, including ethanol and heat, present a backdrop for further engineering, and provide glimpses into the natural variation of stress tolerance in wild yeast strains.
Figures
Fig. 1.—
Stress tolerance profiles. (A) Acquired ethanol tolerance of LEP, MUSH, CRB, and other strains from (Lewis et al. 2010). Cells were exposed to high does of ethanol with (blue) or without (gray) prior treatment of 5% v/v ethanol. (B and C) Growth rates (see Materials and Methods) of S. cerevisiae strains in YPD at 40 °C (B) or in ACSH at 42 °C (C) relative to YPD (adapted from [Sato et al. 2014]).
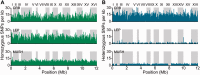
Fig. 2.—
Distribution of SNPs across the genome. Homozygous (A, green) and heterozygous (B, blue) SNPs relative to the S288c reference for 16 S. cerevisiae chromosomes, with a sliding 1-kb window of 100 bp step size.
Fig. 3.—
Population structure of 66 S. cerevisiae strains. Population structure was inferred using 11,795 evenly distributed SNPs and six ancestral populations, identified as European/wine (green), North American/oak (orange), West African (blue), Sake (purple), and Malaysian (yellow) lineages, as well as various human-associated (red) strains. For each strain indicated on the x axis, the height of each colored block represents the proportion of each population assigned to that strain. Labels indicate the source from which each strain was isolated.
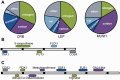
Fig. 4.—
Unique genes in CRB, LEP, and MUSH. A. Functional distribution of 25 non-S288c genes in CRB, 15 non-S288c genes in LEP, and 12 non-S288c genes in MUSH, classified according to predicted GO biological process or molecular function. CW, cell wall; other, unknown or other. (B and C). Genomic architecture of non-S288c genes in CRB and LEP (B) or in CRB and biofuel strain JAY291 (C). Black bars are spaced 2 kb apart.
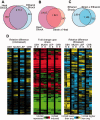
Fig. 5.—
Expression differences across environments and strains. (A) Venn diagram representing the number of differentially expressed genes responding to heat or ethanol stress, regardless of strain background. (B and C) The overlap in heat-responsive genes (independent of strain background, B) or ethanol-responsive genes (independent of strain background, C) and genes with a strain-by-stress interaction. (D) Two hundred forty-eight genes differentially expressed in at least one wild strain. Left: log2 expression differences in unstressed strains versus the average of all strains; Middle: log2-fold changes in each strain responding to heat (H) or ethanol (E) stress, compared with the strain’s starting expression before addition of stress; Right: Difference in stress-responsive expression in each strain versus the average across all strains. Red/green represents increased/decreased expression in response to each stress; yellow/blue represent higher/lower expression in the denoted strain compared with the average of all strains.
Similar articles
- Improved ethanol productivity and ethanol tolerance through genome shuffling of Saccharomyces cerevisiae and Pichia stipitis.
Jetti KD, Gns RR, Garlapati D, Nammi SK. Jetti KD, et al. Int Microbiol. 2019 Jun;22(2):247-254. doi: 10.1007/s10123-018-00044-2. Epub 2018 Nov 15. Int Microbiol. 2019. PMID: 30810988 - Exploiting natural variation in Saccharomyces cerevisiae to identify genes for increased ethanol resistance.
Lewis JA, Elkon IM, McGee MA, Higbee AJ, Gasch AP. Lewis JA, et al. Genetics. 2010 Dec;186(4):1197-205. doi: 10.1534/genetics.110.121871. Epub 2010 Sep 20. Genetics. 2010. PMID: 20855568 Free PMC article. - Ethanol production process driving changes on industrial strains.
Nagamatsu ST, Coutouné N, José J, Fiamenghi MB, Pereira GAG, Oliveira JVC, Carazzolle MF. Nagamatsu ST, et al. FEMS Yeast Res. 2021 Jan 16;21(1):foaa071. doi: 10.1093/femsyr/foaa071. FEMS Yeast Res. 2021. PMID: 33417685 - Stress-related challenges in pentose fermentation to ethanol by the yeast Saccharomyces cerevisiae.
Almeida JR, Runquist D, Sànchez i Nogué V, Lidén G, Gorwa-Grauslund MF. Almeida JR, et al. Biotechnol J. 2011 Mar;6(3):286-99. doi: 10.1002/biot.201000301. Epub 2011 Feb 9. Biotechnol J. 2011. PMID: 21305697 Review. - Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives.
Matsushika A, Inoue H, Kodaki T, Sawayama S. Matsushika A, et al. Appl Microbiol Biotechnol. 2009 Aug;84(1):37-53. doi: 10.1007/s00253-009-2101-x. Epub 2009 Jul 2. Appl Microbiol Biotechnol. 2009. PMID: 19572128 Review.
Cited by
- Efficient engineering of marker-free synthetic allotetraploids of Saccharomyces.
Alexander WG, Peris D, Pfannenstiel BT, Opulente DA, Kuang M, Hittinger CT. Alexander WG, et al. Fungal Genet Biol. 2016 Apr;89:10-17. doi: 10.1016/j.fgb.2015.11.002. Epub 2015 Nov 7. Fungal Genet Biol. 2016. PMID: 26555931 Free PMC article. - Global analysis of mutations driving microevolution of a heterozygous diploid fungal pathogen.
Ene IV, Farrer RA, Hirakawa MP, Agwamba K, Cuomo CA, Bennett RJ. Ene IV, et al. Proc Natl Acad Sci U S A. 2018 Sep 11;115(37):E8688-E8697. doi: 10.1073/pnas.1806002115. Epub 2018 Aug 27. Proc Natl Acad Sci U S A. 2018. PMID: 30150418 Free PMC article. - Into the wild: new yeast genomes from natural environments and new tools for their analysis.
Libkind D, Peris D, Cubillos FA, Steenwyk JL, Opulente DA, Langdon QK, Rokas A, Hittinger CT. Libkind D, et al. FEMS Yeast Res. 2020 Mar 1;20(2):foaa008. doi: 10.1093/femsyr/foaa008. FEMS Yeast Res. 2020. PMID: 32009143 Free PMC article. Review. - Metabolic engineering of Saccharomyces cerevisiae to produce a reduced viscosity oil from lignocellulose.
Tran TNT, Breuer RJ, Avanasi Narasimhan R, Parreiras LS, Zhang Y, Sato TK, Durrett TP. Tran TNT, et al. Biotechnol Biofuels. 2017 Mar 20;10:69. doi: 10.1186/s13068-017-0751-y. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28331545 Free PMC article. - Accurate and Sensitive Quantitation of the Dynamic Heat Shock Proteome Using Tandem Mass Tags.
Storey AJ, Hardman RE, Byrum SD, Mackintosh SG, Edmondson RD, Wahls WP, Tackett AJ, Lewis JA. Storey AJ, et al. J Proteome Res. 2020 Mar 6;19(3):1183-1195. doi: 10.1021/acs.jproteome.9b00704. Epub 2020 Feb 19. J Proteome Res. 2020. PMID: 32027144 Free PMC article.
References
- Aa E, et al. Population structure and gene evolution in Saccharomyces cerevisiae. FEMS Yeast Res. 2006;6:702–715. - PubMed
- Almeida JR, et al. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biotechnol. 2007;82:340.
- Babrzadeh F, et al. Whole-genome sequencing of the efficient industrial fuel-ethanol fermentative Saccharomyces cerevisiae strain CAT-1. Mol Genet Genomics. 2012;287:485–494. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous