Molecular evolution of peste des petits ruminants virus - PubMed (original) (raw)
. 2014 Dec;20(12):2023-33.
doi: 10.3201/eid2012.140684.
Muhammad Munir, AravindhBabu R Parthiban, Ashley C Banyard, Jingyue Bao, Zhiliang Wang, Chrisostom Ayebazibwe, Gelagay Ayelet, Mehdi El Harrak, Mana Mahapatra, Geneviève Libeau, Carrie Batten, Satya Parida
- PMID: 25418782
- PMCID: PMC4257836
- DOI: 10.3201/eid2012.140684
Molecular evolution of peste des petits ruminants virus
Murali Muniraju et al. Emerg Infect Dis. 2014 Dec.
Abstract
Despite safe and efficacious vaccines against peste des petits ruminants virus (PPRV), this virus has emerged as the cause of a highly contagious disease with serious economic consequences for small ruminant agriculture across Asia, the Middle East, and Africa. We used complete and partial genome sequences of all 4 lineages of the virus to investigate evolutionary and epidemiologic dynamics of PPRV. A Bayesian phylogenetic analysis of all PPRV lineages mapped the time to most recent common ancestor and initial divergence of PPRV to a lineage III isolate at the beginning of 20th century. A phylogeographic approach estimated the probability for root location of an ancestral PPRV and individual lineages as being Nigeria for PPRV, Senegal for lineage I, Nigeria/Ghana for lineage II, Sudan for lineage III, and India for lineage IV. Substitution rates are critical parameters for understanding virus evolution because restrictions in genetic variation can lead to lower adaptability and pathogenicity.
Figures
Figure 1
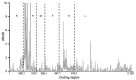
Mean ratios of nonsynonymous (dN) to synonymous (dS) substitutions per site of concatenated coding regions of peste des petits ruminants virus genome. Proportion of dS substitutions per potential dS site and proportion of dN substitutions per potential dN site were calculated by using the method of Nei and Gojobori (29) and the suite of nucleotide analysis program (
). Vertical dashed lines indicate gene junctions with sliding windows of size = 5 codons. dN/dS values ≥ 10 are shown as 10. Numbers along baseline indicate coding regions (basepairs) of individual genes. N, nucleoprotein; P, phosphoprotein; M, matrix; F, fusion; H, hemagglutinin; L, large polymerase.
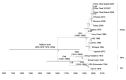
Figure 2
Time-scaled Bayesian maximum clade credibility phylogeny tree based on peste des petits ruminants virus complete genome sequences. The tree was constructed by using the uncorrelated exponential distribution model and exponential tree prior. Branch tips correspond to date of collection and branch lengths reflect elapsed time. Tree nodes were annotated with posterior probability values and estimated median dates of time to most recent common ancestor (TMRCA). Corresponding 95% highest posterior density (HPD) interval values of TMRCA are indicated as gray bars. Horizontal axis indicates time in years. UAE, United Arab Emirates.
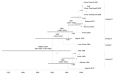
Figure 3
Time-scaled Bayesian MCC phylogeny tree based on peste des petits ruminants virus (PPRV), rinderpest virus (RPV), and measles virus (MV) complete genome sequences. The tree was constructed by using the uncorrelated exponential distribution model and exponential tree prior. Branch tips correspond to date of collection and branch lengths reflect elapsed time. Tree nodes were annotated with posterior probability values, estimated median dates of time to most recent common ancestor (TMRCA). Corresponding 95% highest posterior density (HPD) values of TMRCA are indicated as gray bars. Horizontal axis indicates time in years. UAE, United Arab Emirates.
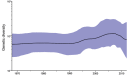
Figure 4
Bayesian skyline plot showing demographic history of global peste des petits ruminants viruses sampled during 1968–2012. Genetic diversity was estimated by using a partial nucleoprotein gene dataset (n = 159). The thick black line represents median genetic diversity and the blue shaded areas show 95% highest posterior density estimate.
Figure 5
Maximum clade credibility tree constructed for the geospatial analysis of peste des petits ruminants viruses by using complete genome data. Nodes are colored according to the most probable location of their ascendent locations. Posterior probability values are shown along tree nodes. Posterior probability distribution (PPD) values of root location states of the ancestral node are shown along the x-axis at the top left. UAE, United Arab Emirates.
Figure 6
Probability of root locations of the most recent common ancestral peste des petits ruminants (PPRV). MCC trees were obtained by using the continuous time Markov chain and Bayesian stochastic search variable selection procedures. Root location probabilities of the most recent common ancestor using global PPRV isolates (panel A ) are shown graphically alongside lineages I–IV (panels B–E) and were estimated by using a complete dataset of PPRV partial nucleoprotein gene data and individual lineages separately. Probabilities of root locations are shown as percentages along the x-axes. UAE, United Arab Emirates; CAR, Central African Republic.
Similar articles
- Genome sequencing of an Indian peste des petits ruminants virus isolate, Izatnagar/94, and its implications for virus diversity, divergence and phylogeography.
Sahu AR, Wani SA, Saminathan M, Rajak KK, Sahoo AP, Pandey A, Saxena S, Kanchan S, Tiwari AK, Mishra B, Muthuchelvan D, Singh RP, Singh Y, Baig M, Mishra BP, Singh RK, Gandham RK. Sahu AR, et al. Arch Virol. 2017 Jun;162(6):1677-1693. doi: 10.1007/s00705-017-3288-2. Epub 2017 Feb 28. Arch Virol. 2017. PMID: 28247095 - Evolutionary dynamics of recent peste des petits ruminants virus epidemic in China during 2013-2014.
Bao J, Wang Q, Li L, Liu C, Zhang Z, Li J, Wang S, Wu X, Wang Z. Bao J, et al. Virology. 2017 Oct;510:156-164. doi: 10.1016/j.virol.2017.07.018. Epub 2017 Jul 19. Virology. 2017. PMID: 28734191 Free PMC article. - Emergence of Lineage IV Peste des Petits Ruminants Virus in Ethiopia: Complete Genome Sequence of an Ethiopian Isolate 2010.
Muniraju M, Mahapatra M, Ayelet G, Babu A, Olivier G, Munir M, Libeau G, Batten C, Banyard AC, Parida S. Muniraju M, et al. Transbound Emerg Dis. 2016 Aug;63(4):435-42. doi: 10.1111/tbed.12287. Epub 2014 Nov 14. Transbound Emerg Dis. 2016. PMID: 25400010 - Peste des Petits Ruminants Virus.
Baron MD, Diallo A, Lancelot R, Libeau G. Baron MD, et al. Adv Virus Res. 2016;95:1-42. doi: 10.1016/bs.aivir.2016.02.001. Epub 2016 Mar 14. Adv Virus Res. 2016. PMID: 27112279 Review. - Peste Des Petits Ruminants in Benin: Persistence of a Single Virus Genotype in the Country for Over 42 Years.
Adombi CM, Waqas A, Dundon WG, Li S, Daojin Y, Kakpo L, Aplogan GL, Diop M, Lo MM, Silber R, Loitsch A, Diallo A. Adombi CM, et al. Transbound Emerg Dis. 2017 Aug;64(4):1037-1044. doi: 10.1111/tbed.12471. Epub 2016 Jan 22. Transbound Emerg Dis. 2017. PMID: 26801518 Review.
Cited by
- Transcriptional Profiles of Murine Bone Marrow-Derived Dendritic Cells in Response to Peste des Petits Ruminants Virus.
Li L, Wu J, Liu D, Du G, Liu Y, Shang Y, Liu X. Li L, et al. Vet Sci. 2019 Nov 29;6(4):95. doi: 10.3390/vetsci6040095. Vet Sci. 2019. PMID: 31795377 Free PMC article. - Morbillivirus: A highly adaptable viral genus.
Libbey JE, Fujinami RS. Libbey JE, et al. Heliyon. 2023 Jul 7;9(7):e18095. doi: 10.1016/j.heliyon.2023.e18095. eCollection 2023 Jul. Heliyon. 2023. PMID: 37483821 Free PMC article. Review. - A comparative phylogenomic analysis of peste des petits ruminants virus isolated from wild and unusual hosts.
Rahman AU, Munir M, Shabbir MZ. Rahman AU, et al. Mol Biol Rep. 2019 Oct;46(5):5587-5593. doi: 10.1007/s11033-019-04973-7. Epub 2019 Jul 17. Mol Biol Rep. 2019. PMID: 31317455 - Peste des Petits Ruminants at the Wildlife-Livestock Interface in the Northern Albertine Rift and Nile Basin, East Africa.
Fernandez Aguilar X, Mahapatra M, Begovoeva M, Kalema-Zikusoka G, Driciru M, Ayebazibwe C, Adwok DS, Kock M, Lukusa JK, Muro J, Marco I, Colom-Cadena A, Espunyes J, Meunier N, Cabezón O, Caron A, Bataille A, Libeau G, Parekh K, Parida S, Kock R. Fernandez Aguilar X, et al. Viruses. 2020 Mar 7;12(3):293. doi: 10.3390/v12030293. Viruses. 2020. PMID: 32156067 Free PMC article. - Peste des petits ruminants.
Parida S, Muniraju M, Mahapatra M, Muthuchelvan D, Buczkowski H, Banyard AC. Parida S, et al. Vet Microbiol. 2015 Dec 14;181(1-2):90-106. doi: 10.1016/j.vetmic.2015.08.009. Epub 2015 Sep 5. Vet Microbiol. 2015. PMID: 26443889 Free PMC article. Review.
References
- de Swart RL, Duprex WP, Osterhaus AD. Rinderpest eradication: lessons for measles eradication? Curr Opin Virol. 2012;2:330–4. - PubMed
Publication types
MeSH terms
Grants and funding
- BB/H009485/1/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L004801/1/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L013657/1/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources