ROR nuclear receptors: structures, related diseases, and drug discovery - PubMed (original) (raw)
Review
ROR nuclear receptors: structures, related diseases, and drug discovery
Yan Zhang et al. Acta Pharmacol Sin. 2015 Jan.
Erratum in
- Correction.
[No authors listed] [No authors listed] Acta Pharmacol Sin. 2015 Feb;36(2):290. doi: 10.1038/aps.2015.2. Acta Pharmacol Sin. 2015. PMID: 25645678 Free PMC article. No abstract available.
Abstract
Nuclear receptors (NRs) are ligand-regulated transcription factors that regulate metabolism, development and immunity. The NR superfamily is one of the major classes of drug targets for human diseases. Retinoic acid receptor-related orphan receptor (ROR) α, β and γ belong to the NR superfamily, and these receptors are still considered as 'orphan' receptors because the identification of their endogenous ligands has been controversial. Recent studies have demonstrated that these receptors are regulated by synthetic ligands, thus emerge as important drug targets for the treatment of multiple sclerosis, rheumatoid arthritis, psoriasis, etc. Studying the structural basis and ligand development of RORs will pave the way for a better understanding of the roles of these receptors in human diseases. Here, we review the structural basis, disease relevance, strategies for ligand identification, and current status of development of therapeutic ligands for RORs.
Figures
Figure 1
Structural organization of ROR functional domains. (A) Schematic diagram of the domain structure of RORs. Similar to other NRs, RORs display conserved modular domain architecture with a N-terminal ligand-independent activation function 1 (AF-1) domain, followed by a DNA binding domain (DBD), a hinge domain, and a ligand-binding domain with an activation function 2 (AF-2) domain. The DBD binds specific DNA sequences that typically consist of TAAA/TNTA_GGTCA_ (termed ROR response element, RORE). (B) Sequence alignment of the ligand binding domain of RORα, RORβ, and RORγ performed using ClustalW. Cartoon presentation of the general architecture of RORs was shown under the corresponding sequences. Identical residues are labeled with an asterisk. Partially conserved residues are labeled with a colon. The residue numbering for RORα, RORβ, and RORγ are E305-G556, E222-K470, and E269-K518, respectively. Residues around the ligand are shown as red letters. Residues important for ligand binding were labeled on top of the sequences.
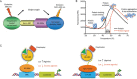
Figure 2
Structural model of ROR agonism and antagonism. (A) RORγ agonists, such as 25-hydroxycholesterol, drive recruitment of transcriptional coactivators, which leads to the modulation and promotion of target gene transcription. Inverse agonists of RORγ, such as digoxin, disrupt recruitment of the transcriptional coactivator and repress target gene expression. (B) Agonist binding induces a conformational change and facilitates binding of the LXXLL motif of coactivators, such as SRC2. Antagonists, such as digoxin, induce a conformational change of helix 12 and circumvent the coactivator recruitment. The coactivator protein and helix 12 are colored in red and green, respectively. The agonist (left, 3L0L.pdb) and inverse agonist (right, 3B0W.pdb) are shown as sticks.
Figure 3
Comparison of the binding of 25-hydroxycholesterol (A), digoxin (B), T0901317 (C), and GSK-2 (D) in the RORγ ligand-binding pocket. Hydrogen bonds are depicted as dashed lines (red). (A) 25-OHC (green) formed direct hydrogen bonds with Gln286 and His479, and water-mediated hydrogen bonds with Arg364, Arg367, and Tyr502. (B) Digoxin (cyan) formed direct hydrogen bonds with Arg367, Phe377, His479, and Leu391, and water-mediated hydrogen bonds with Val361 and Glu379. (C) The phenylsulfonamide group of T0901317 (purple) forms a π−π stacking interaction with Phe378 and Phe388. The ligand and His479 disrupt the interaction network originally formed with Trp317, Tyr502, and Phe506. (D) GSK-2 (orange) formed direct hydrogen bonds with Leu287, Arg367, and Phe377. The amide carbonyl of GSK-2 formed a water-mediated hydrogen bond with Gln286 and His323. The aniline ring of GSK-2 also formed a π−π stacking interaction with Phe378 and Phe388.
Figure 4
Representative drug discovery strategies. (A) Schematic representation of the AlphaScreen assay (Amplified Luminescent Proximity Homogenous Screen Assay Screen). H6-RORγ is immobilized on Ni-chelating acceptor beads and the biotinylated coactivator (Biotin-SRC) on streptavidin-coated donor beads. Donor beads contain a photosensitizer that, upon activation at 680 nm, converts ambient oxygen to singlet oxygen. If the acceptor beads are brought into close proximity of the donor beads by a RORγ-coactivator interaction, energy is transferred from the singlet oxygen to the thioxene derivatives in the acceptor beads, which results in light emission at 520–620 nm. Addition of a RORγ inverse agonist represses the signal of acceptor bead-immobilized H6-RORγ and donor-bead-biotin-coactivator. (B) Schematic representation of fluorescence intensity versus temperature of the melting protein in the presence of SYPRO orange. Ligand binding to a target protein can stabilize a protein's native state reflected in the increased melting temperature (Tm) of the bound protein. Monitoring of the ΔTm of apo and ligand-bound proteins can be used to determine the ligand binding affinity. (C) Cell-based reporter assays. [left] Ligand binds to the NR LBD and the NR DBD binds to the nuclear receptor response element upstream of the reporter gene to activate transcription. [right] Upon ligand binding, the GAL4-NR-LBD binds to the GAL4 UAS to activate transcription.
Figure 5
Structures of RORγ inverse agonists from scripps institute (A), GSK (B), and Japan tobacco (C).
Similar articles
- Ligand regulation of retinoic acid receptor-related orphan receptors: implications for development of novel therapeutics.
Solt LA, Griffin PR, Burris TP. Solt LA, et al. Curr Opin Lipidol. 2010 Jun;21(3):204-11. doi: 10.1097/MOL.0b013e328338ca18. Curr Opin Lipidol. 2010. PMID: 20463469 Free PMC article. Review. - Structure-activity relationship-guided development of retinoic acid receptor-related orphan receptor gamma (RORγ)-selective inverse agonists with a phenanthridin-6(5H)-one skeleton from a liver X receptor ligand.
Nishiyama Y, Nakamura M, Misawa T, Nakagomi M, Makishima M, Ishikawa M, Hashimoto Y. Nishiyama Y, et al. Bioorg Med Chem. 2014 May 1;22(9):2799-808. doi: 10.1016/j.bmc.2014.03.007. Epub 2014 Mar 17. Bioorg Med Chem. 2014. PMID: 24702856 - Orphan nuclear receptors and the regulation of nutrient metabolism: understanding obesity.
Pearen MA, Muscat GE. Pearen MA, et al. Physiology (Bethesda). 2012 Jun;27(3):156-66. doi: 10.1152/physiol.00007.2012. Physiology (Bethesda). 2012. PMID: 22689791 Review. - The emerging roles of orphan nuclear receptors in prostate cancer.
Wu D, Cheung A, Wang Y, Yu S, Chan FL. Wu D, et al. Biochim Biophys Acta. 2016 Aug;1866(1):23-36. doi: 10.1016/j.bbcan.2016.06.001. Epub 2016 Jun 2. Biochim Biophys Acta. 2016. PMID: 27264242 Review. - Natural products as modulators of retinoic acid receptor-related orphan receptors (RORs).
Ladurner A, Schwarz PF, Dirsch VM. Ladurner A, et al. Nat Prod Rep. 2021 Apr 28;38(4):757-781. doi: 10.1039/d0np00047g. Nat Prod Rep. 2021. PMID: 33118578 Review.
Cited by
- Discovery of a Novel Class of Dual GPBAR1 Agonists-RORγt Inverse Agonists for the Treatment of IL-17-Mediated Disorders.
Fiorillo B, Roselli R, Finamore C, Biagioli M, di Giorgio C, Bordoni M, Conflitti P, Marchianò S, Bellini R, Rapacciuolo P, Cassiano C, Limongelli V, Sepe V, Catalanotti B, Fiorucci S, Zampella A. Fiorillo B, et al. ACS Omega. 2023 Jan 31;8(6):5983-5994. doi: 10.1021/acsomega.2c07907. eCollection 2023 Feb 14. ACS Omega. 2023. PMID: 36816679 Free PMC article. - Discovery of a Series of Pyrazinone RORγ Antagonists and Identification of the Clinical Candidate BI 730357.
Harcken C, Csengery J, Turner M, Wu L, Liang S, Sibley R, Brunette S, Labadia M, Hoyt K, Wayne A, Wieckowski T, Davis G, Panzenbeck M, Souza D, Kugler S, Terenzio D, Collin D, Smith D, Fryer RM, Tseng YC, Hehn JP, Fletcher K, Hughes RO. Harcken C, et al. ACS Med Chem Lett. 2021 Jan 5;12(1):143-154. doi: 10.1021/acsmedchemlett.0c00575. eCollection 2021 Jan 14. ACS Med Chem Lett. 2021. PMID: 33488976 Free PMC article. - Structural determinant for inducing RORgamma specific inverse agonism triggered by a synthetic benzoxazinone ligand.
Marcotte DJ, Liu Y, Little K, Jones JH, Powell NA, Wildes CP, Silvian LF, Chodaparambil JV. Marcotte DJ, et al. BMC Struct Biol. 2016 Jun 1;16(1):7. doi: 10.1186/s12900-016-0059-3. BMC Struct Biol. 2016. PMID: 27246200 Free PMC article. - Th17 Cells in Viral Infections-Friend or Foe?
Paiva IA, Badolato-Corrêa J, Familiar-Macedo D, de-Oliveira-Pinto LM. Paiva IA, et al. Cells. 2021 May 11;10(5):1159. doi: 10.3390/cells10051159. Cells. 2021. PMID: 34064728 Free PMC article. Review. - Retinoid-related orphan nuclear receptor alpha (RORα)-deficient mice display morphological testicular defects.
Sayed RKA, Mokhtar DM, Fernández-Ortiz M, Escames G, Acuña-Castroviejo D. Sayed RKA, et al. Lab Invest. 2019 Dec;99(12):1835-1849. doi: 10.1038/s41374-019-0299-5. Epub 2019 Aug 13. Lab Invest. 2019. PMID: 31409890
References
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell 1995; 83: 841–50. - PubMed
- Giguere V. Orphan nuclear receptors: from gene to function. Endocr Rev 1999; 20: 689–725. - PubMed
- Laudet V. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J Mol Endocrinol 1997; 19: 207–26. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases