Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14 - PubMed (original) (raw)
Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14
Yong-Qiang Ning et al. Nucleic Acids Res. 2015.
Abstract
The histone demethylase JMJ14 catalyzes histone demethylation at lysine 4 of histone 3 and is involved in transcriptional repression and flowering time control in Arabidopsis. Here, we report that JMJ14 is physically associated with two previously uncharacterized NAC transcription factors, NAC050 and NAC052. The NAC050/052-RNAi plants and the CRISPR-CAS9-mediated nac050/052 double mutant plants show an early flowering phenotype, which is similar to the phenotype of jmj14, suggesting a functional association between JMJ14 and NAC050/052. RNA-seq data indicated that hundreds of common target genes are co-regulated by JMJ14 and NAC50/052. Our ChIP analysis demonstrated that JMJ14 and NAC050 directly bind to co-upregulated genes shared in jmj14 and NAC050/052-RNAi, thereby facilitating H3K4 demethylation and transcriptional repression. The NAC050/052 recognition DNA cis-element was identified by an electrophoretic mobility shift assay at the promoters of its target genes. Together, our study identifies two novel NAC transcription repressors and demonstrates that they are involved in transcriptional repression and flowering time control by associating with the histone demethylase JMJ14.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
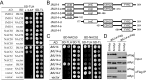
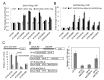
Figure 1.
JMJ14 associates with NAC050 and NAC052 in vivo. (A) The NAC transcription factors NAC050 and NAC052 were identified in proteins that were co-purified with JMJ14 in a mass spectrometric assay. The protein extract was isolated from flowers of 1-month-old plants. (B) The interaction of JMJ14 with NAC050 and NAC052 was determined by co-IP. The JMJ14–3xFlag transgenic plants were crossed with NAC050–3xMyc and NAC052–3xMyc transgenic plants. The offspring harboring both of the epitope tags was used in the co-IP analysis. The parent JMJ14–3xFlag, NAC050–3xMyc and NAC052–3xMyc transgenic plants were used as controls. Two-week-old seedlings were used for protein extract. (C) Gel filtration assay of the protein extracts from JMJ14–3xFlag, NAC050–3xMyc and NAC052–3xMyc transgenic plants. The protein extract was isolated from flowers of 1-month-old plants.
Figure 2.
The FYRC domain is required for the interaction of JMJ14 with NAC050 and NAC052. (A) The interaction of JMJ14 with NAC050 and NAC052 was determined by the yeast two-hybrid assay. The full-length cDNA sequences of JMJ14, NAC050 and NAC052 were cloned into the yeast vectors pGADT7 and pGBKT7. The constructs were transformed into the yeast strain Y1348 as indicated and then subjected to a growth assay on SD-TLH (synthetic dropout medium minus Trp, Leu and His) supplemented with 20 mM 3-AT as well as on SD-TL. (B) Diagram of the full-length and truncated versions of JMJ14. The conserved domains JmjN, JmjC, C5HC2 and FYRC are indicated. (C) Different JMJ14 cDNA fragments were cloned into pGADT7. The constructs harboring each of the JMJ14 sequences, as well as pGBKT7-NAC050 or pGBKT7-NAC052, were co-transformed into the yeast strain Y1348 for growth assays. (D) Either the full-length JMJ14–3xFlag or the truncated JMJ14-a-3xFlag transgene was introduced into NAC-050–3xMyc or NAC052–3xMyc transgenic plants. The transgenic plants were used to determine the interaction of the full-length and truncated JMJ14 with NAC050 or NAC052 by co-IP. The protein extract for co-IP was isolated from 2-week-old seedlings.
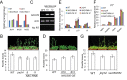
Figure 3.
Functional association between JMJ14 and NAC050/052 in flowering time regulation. (A) Quantitative RT-PCR was performed to determine the transcript levels of JMJ14, NAC050 and NAC052 in the WT, jmj14 and two independent NAC050/052 knockdown lines, NAC-RNAi-8 and NAC-RNAi-11. Total RNA used for RT-PCR was extracted from 2-week-old seedlings. The actin gene was used as an internal control. The results of three replicates are shown. (B) Shown are the WT, jmj14, NAC-RNAi-8 and NAC-RNAi-11 plants that were grown on soil under long-day conditions. The flowering time was assessed by counting rosette leaves. At least 20 plants of each genotype were used to count the rosette leaves. The average and standard deviation are indicated in the chart. (C) The transcript level of NAC050 was determined by RT-PCR in the WT and two individual 35S-NAC050 transgenic lines #10 and #12. The actin gene was used as a control. ‘No RT’ stands for amplification of the actin gene without reverse transcription. (D) The WT and 35S-NAC050 transgenic plants were grown on soil under long-day conditions. The numbers of rosette leaves from at least 20 plants were included to calculate the average and standard deviation. (E) The effect of jmj14 and NAC050/052-RNAi on the expression of floral integrator genes. The expression of the floral integrator genes FT, LFY and PI in the WT, jmj14 and two individual NAC050/052-RNAi lines was determined by quantitative RT-PCR. The experiment was biologically repeated and the results of three technical replicates from a representative experiment are shown. (F) The effect of jmj14 and NAC050/052-RNAi on the expression of FT in different tissues of 1-month-old plants. The results of three replicates are shown. (G) The CRISPR-mediated nac050/052 double mutant shows an early flowering phenotype. Soil-grown plants were photographed 3 weeks after planting.
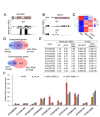
Figure 4.
Effect of jmj14 and NAC050/052-RNAi on gene expression as determined by RNA-seq. (A) The plots show the RNA reads that matched NAC050 and NAC052. The RNA reads were plotted in the WT and NAC050/052-RNAi plants. (B) The JMJ14 RNA reads were plotted in the WT and jmj14 plants. RNA reads of a flanking gene are comparable between the WT and jmj14 plants and are shown as a control. (C) Heat map of differentially expressed genes in jmj14 and NAC-RNAi plants relative to the WT. (D) Venn diagram showing the overlap of differentially expressed genes in jmj14 and NAC-RNAi plants. (E) Showing is the list of co-upregulated genes that were selected for quantitative RT-PCR assay. Normalized reads in each ecotype and _P-_values are indicated. (F) Quantitative RT-PCR was performed to confirm the effect of jmj14 and NAC050/052-RNAi on the expression of their target genes as determined by RNA-seq. Two individual NAC050/052-RNAi lines, 8 and 11, were used. The expression of the actin gene was used as an internal control. The results of three technical replicates were shown. Total RNA used for RNA-seq and quantitative RT-PCR was extracted from 2-week-old seedlings.
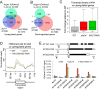
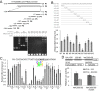
Figure 5.
Effect of jmj14 and NAC050/052-RNAi on H3K4me3 as determined by ChIP-seq. (A and B) Venn diagrams showing the overlap between the H3K4me3 hypermethylated genes in jmj14 and the upregulated (A) or downregulated genes (B) in jmj14 and NAC050/052-RNAi plants. (C) Box plot showing the effect of jmj14 and NAC050/052-RNAi on gene expression for 494 co-upregulated genes in jmj14 and nac050/052-RNAi. (D) H3K4me3 of the 494 overlapping upregulated genes in jmj14 and NAC050/052-RNAi plants was plotted for the transcription regions along with the 1-kb upstream and downstream flanking regions. The _y_-axis indicates the normalized reads number. (E) H3K4me3 hypermethylated genes identified by ChIP-seq were confirmed by ChIP-PCR in the WT, jmj14 and NAC050/052-RNAi plants. Diagrams show positions of all DNA fragments amplified in the ChIP-PCR assay. The hypermethylated sites include AT1G11540-A, AT2G18193-A and AT5G27940-A. The sites that are adjacent to the H3K4me3 hypermethylated regions were used as negative controls. These sites are AT1G11540-B, AT2G18193-B, AT5G27940-B and AT5G27940-C. Two-week-old seedlings were used for ChIP-seq and ChIP-PCR.
Figure 6.
JMJ14 and NAC050 bind to their common target genes. (A) JMJ14-Flag ChIP-PCR was performed to determine the occupancy of JMJ14-Flag at the common targets of JMJ14 and NAC050/052. The JMJ14–3xFlag construct was transformed and stably expressed in the WT as well as in NAC050/052-RNAi plants. (B) NAC050-Myc ChIP-PCR was performed to examine the occupancy of NAC050–3xMyc at the common targets of JMJ14 and NAC050/052. The NAC050–3xMyc construct was introduced into the WT and jmj14 plants. The expression levels of NAC050–3xMyc in the WT and jmj14 plants are equivalent. Two-week-old seedlings of each genotype were used in the ChIP-PCR assay for JMJ14-Flag and NAC-Myc. (C) The transcriptional repression activity of NAC050 was determined by the transient expression of the luciferase reporter in protoplast cells. The luciferase reporter gene was driven by the promoter sequences of AT2G18720, AT2G21640, AT5G16020 and AT1G02580. The 35S-NAC050 construct was transformed to determine whether the overexpression of NAC050 represses the luciferase activity. (D) The luciferase reporter gene was driven by the minimal 35S promoter with the upstream GAL4-binding site. In the effector construct, the transcriptional activator VP16 fused to the GAL4 DNA-binding domain activates the transcription of the reporter gene. Either the full-length NAC050 or truncated NAC sequences were ligated in frame with the GAL4-VP16 fusion sequence in the effector construct. The reporter construct and each of the effector constructs were co-transformed into protoplast cells for the luciferase activity assay. The experiments in this figure were biologically repeated and the same results were obtained. Showing is the results of three technical replicates from a representative experiment.
Figure 7.
Identification and characterization of the NAC050/052-binding DNA cis-element. (A) NAC050 binds to the promoter sequence of its target gene UGT74E2 as determined by EMSA. A series of DNA fragments in the AT1G05680 promoter were used in the binding assay. (B) The oligonucleotide sequence AT1G05680-P9 harboring the NAC050 binding site was mutated at a series of three nucleotides and then used in the NAC050 binding assay. The binding levels were normalized by the binding between NAC050 and the WT oligonucleotide sequence. Shown are the results from three independent replicates. (C) The AT1G05680-P9 oligonucleotide was subjected to mutation at one residue and was then used in the NAC050 binding assay by EMSA. Each nucleotide was mutated to three other nucleotides to determine the alternation of the nucleotide. (D) The luciferase reporter gene was driven by the 35S minimal promoter ligated with GAL4 binding element in front of a 5xNAC050-binding element (CTTGGTCGC
CACG
GAA). The 5xNAC050-binding element was substituted with CTTGGTCGC
CCCG
GAA and used as a negative control. The expression of the reporter gene was determined by the luciferase activity assay in protoplast cells. The effector construct GAL4-DBD-VP16 was used to activate the reporter gene. The NAC050 expression construct was transformed to determine the effect of NAC050 on the expression of the reporter gene.
Similar articles
- C-terminal domains of a histone demethylase interact with a pair of transcription factors and mediate specific chromatin association.
Zhang S, Zhou B, Kang Y, Cui X, Liu A, Deleris A, Greenberg MV, Cui X, Qiu Q, Lu F, Wohlschlegel JA, Jacobsen SE, Cao X. Zhang S, et al. Cell Discov. 2015;1:15003-. doi: 10.1038/celldisc.2015.3. Epub 2015 Apr 28. Cell Discov. 2015. PMID: 26617990 Free PMC article. - Metabolic control of histone demethylase activity involved in plant response to high temperature.
Cui X, Zheng Y, Lu Y, Issakidis-Bourguet E, Zhou DX. Cui X, et al. Plant Physiol. 2021 Apr 23;185(4):1813-1828. doi: 10.1093/plphys/kiab020. Plant Physiol. 2021. PMID: 33793949 Free PMC article. - Histone demethylase PHF8 regulates hypoxia signaling through HIF1α and H3K4me3.
Maina PK, Shao P, Jia X, Liu Q, Umesalma S, Marin M, Long D Jr, Concepción-Román S, Qi HH. Maina PK, et al. Biochim Biophys Acta Gene Regul Mech. 2017 Sep;1860(9):1002-1012. doi: 10.1016/j.bbagrm.2017.07.005. Epub 2017 Jul 20. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 28734980 Free PMC article. - Control of the transition to flowering by chromatin modifications.
He Y. He Y. Mol Plant. 2009 Jul;2(4):554-564. doi: 10.1093/mp/ssp005. Epub 2009 Mar 5. Mol Plant. 2009. PMID: 19825638 Review. - Histone demethylases and control of gene expression in plants.
Prakash S, Singh R, Lodhi N. Prakash S, et al. Cell Mol Biol (Noisy-le-grand). 2014 Dec 24;60(5):97-105. Cell Mol Biol (Noisy-le-grand). 2014. PMID: 25535719 Review.
Cited by
- A novel strategy to map a locus associated with flowering time in canola (Brassica napus L.).
Long Y, Zheng P, Anderson JV, Horvath DP, Sthapit J, Li X, Rahman M, Chao WS. Long Y, et al. Mol Genet Genomics. 2024 Oct 8;299(1):95. doi: 10.1007/s00438-024-02191-w. Mol Genet Genomics. 2024. PMID: 39379673 Free PMC article. - Genome-Wide Identification of NAC Family Genes and Their Expression Analyses in Response to Osmotic Stress in Cannabis sativa L.
Li Q, Zhang H, Yang Y, Tang K, Yang Y, Ouyang W, Du G. Li Q, et al. Int J Mol Sci. 2024 Aug 30;25(17):9466. doi: 10.3390/ijms25179466. Int J Mol Sci. 2024. PMID: 39273412 Free PMC article. - AdNAC20 Regulates Lignin and Coumarin Biosynthesis in the Roots of Angelica dahurica var. Formosana.
Qu W, Huang W, Chen C, Chen J, Zhao L, Jiang Y, Du X, Liu R, Chen Y, Hou K, Xu D, Wu W. Qu W, et al. Int J Mol Sci. 2024 Jul 22;25(14):7998. doi: 10.3390/ijms25147998. Int J Mol Sci. 2024. PMID: 39063240 Free PMC article. - The H3K4 demethylase JMJ1 is required for proper timing of flowering in Brachypodium distachyon.
Liu B, Li C, Li X, Wang J, Xie W, Woods DP, Li W, Zhu X, Yang S, Dong A, Amasino RM. Liu B, et al. Plant Cell. 2024 Jul 2;36(7):2729-2745. doi: 10.1093/plcell/koae124. Plant Cell. 2024. PMID: 38652680 - Comprehensive Analyses of Four PhNF-YC Genes from Petunia hybrida and Impacts on Flowering Time.
Bin J, Tan Q, Wen S, Huang L, Wang H, Imtiaz M, Zhang Z, Guo H, Xie L, Zeng R, Wei Q. Bin J, et al. Plants (Basel). 2024 Mar 6;13(5):742. doi: 10.3390/plants13050742. Plants (Basel). 2024. PMID: 38475587 Free PMC article.
References
- Baumbusch L.O., Thorstensen T., Krauss V., Fischer A., Naumann K., Assalkhou R., Schulz I., Reuter G., Aalen R.B. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. - PMC - PubMed
- Springer N.M., Napoli C.A., Selinger D.A., Pandey R., Cone K.C., Chandler V.L., Kaeppler H.F., Kaeppler S.M. Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 2003;132:907–925. - PMC - PubMed
- Tsukada Y., Fang J., Erdjument-Bromage H., Warren M.E., Borchers C.H., Tempst P., Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. - PubMed
- Pedersen M.T., Helin K. Histone demethylases in development and disease. Trends Cell Biol. 2010;20:662–671. - PubMed
- Lu F., Li G., Cui X., Liu C., Wang X.J., Cao X. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. Jo. Integr. Plant Biol. 2008;50:886–896. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases