miR-146a is directly regulated by STAT3 in human hepatocellular carcinoma cells and involved in anti-tumor immune suppression - PubMed (original) (raw)
miR-146a is directly regulated by STAT3 in human hepatocellular carcinoma cells and involved in anti-tumor immune suppression
Xiaoxia Sun et al. Cell Cycle. 2015.
Abstract
MicroRNAs (miRNAs) play an important role in tumorigenesis, but their role in tumor-induced immune suppression is largely unknown. STAT3 signaling, a key pathway mediating immune suppression in the tumor microenvironment, is responsible for the transcription of several important miRNAs. In this study, we observed that miR-146a, a known important regulator of immune responses, was downregulated by blocking activated STAT3 in hepatocellular carcinoma (HCC) cells. Furthermore, miR-146a inhibition in HCC cells not only altered the STAT3 activation-associated cytokine profile but also reversed HCC-induced NK cell dysfunction in vitro and improved the anti-tumor effect of lymphocytes in vivo. Importantly, ChIP and luciferase reporter assays confirmed that STAT3 directly bound to the miR-146a promoter and induced miR-146a expression. These findings indicated that miR-146a expression was regulated by aberrantly activated STAT3 in HCC cells and exerted negative effects on anti-tumor immune response, which resulted in the upregulation of cytokines such as TGF-β, IL-17, VEGF and downregulation of type I IFN to create an immunosuppressive microenvironment. This further insight into understanding the mechanism responsible for tumor-induced immune suppression highlights the potential application of miR-146a as a novel immunotherapeutic target for HCC.
Keywords: HCC; STAT3; anti-tumor immune suppression; miR-146a; miRNA.
Figures
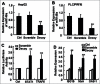
Figure 1.
The expression level of miR-146a in human HCC cells was inhibited by blocking STAT3. After transfecting STAT3 decoy ODN (Decoy), scramble ODN (Scramble), or Lipofectamine reagent control (Ctrl) into HepG2 (A) and PLC/PRF/5 (B) cells for 24 h, miR-146a expression was measured by qPCR analysis. (C) Luciferase activity of pmiR-Reporter vector containing the mRNA 3′-UTR miR-146a target genes STAT1 and TRAF6 was detected using a dual-GloTM Luciferase assay system. (D) Levels of ISG15, MxA, and OAS-1 mRNA were analyzed by qPCR analysis. Data are representative of 3 independent experiments, and statistical significance was determined as **P < 0.01 and *P < 0.05 compared to Lipo-Ctrl.
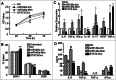
Figure 2.
miR-146a promoted the expression of inflammatory cytokines associated with STAT3 activation in HCC cells. As described in the Materials and Methods section, HepG2 cells were transfected with negative control RNA (NC), miR-146a mimics (miR146a-Mim), miR-146a inhibitors (miR146a-Inh), or STAT3 decoy ODN (STAT3-Dec). (A) HepG2 proliferation was analyzed by MTT assay at the indicated time points. (B) Cell cycle was determined by flow cytometry. The levels of inflammatory cytokines associated with STAT3 activation were determined by qPCR (C) and ELISA (D) analysis. Data are representative of 3 independent experiments, and statistical significance was determined as **P < 0.01 and *P < 0.05 compared to NC.
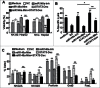
Figure 3.
STAT3 directly regulated miR-146a expression in HCC. (A) A ChIP assay was performed 24 h after STAT3 decoy ODN (Dec) or scramble ODN (Scr) treatment to evaluate the recruitment of STAT3 on miR-146a promoter (miR146a-pro). (B) The level of phosphorylated STAT3 (p-STAT3) (Tyr705) in IL-6–stimulated (400 U/mL) HepG2 cells was examined by western blotting (upper), and a qPCR assay was used to detect the expression of miR-146a in HepG2 cells (lower). (C) After IL-6 stimulation, a ChIP-PCR assay was performed using an anti–p-STAT3705 antibody or rabbit IgG as a control. (D) The luciferase activity of the miR-146a promoter (miR146a-pro) in HepG2 cells was measured using a dual-GloTM Luciferase assay system. The ratio of firefly to Renilla luciferase activity with pGL3-TK-Luciferase transfection was set as 1. Data are representative of 3 independent experiments, and statistical significance was determined as **P < 0.01 and *P < 0.05 compared to control.
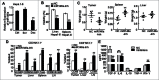
Figure 4.
miR-146a contributed to human HCC–induced immune suppression in vitro. After HepG2 cells were transfected with negative control RNA (NC), miR-146a mimics (miR146a-Mim), miR-146a inhibitors (miR146a-Inh), or STAT3 decoy ODN (STAT3-Dec) as described in the Materials and Methods section, supernatant was collected and incubated with NK-92 or NKL cells in an _in vitro_culture for 12 h. (A) The inhibitory effect of these NK cells on the viability of naïve HepG2 cells was then analyzed by an MTT assay at an E:T ratio of 5:1. (B) The specific lysis of HepG2 cells by these NK-92 cells was detected using a 4-h CFSE/7-AAD flow cytometry assay at an E:T ratio of 5:1. (C) The molecules associated with NK cell cytolysis were examined by flow cytometry. The data in the histograms represent the statistical analysis of the percentage of positive cells. Medium, NK cells cultured in α-MEM alone without any supernatant from HCC cells. Data are representative of 3 independent experiments, and statistical significance was determined as **P < 0.01 and *P < 0.05 compared to NC.
Figure 5.
Inhibition of miR-146a promoted the anti-tumor immune response in vivo. (A) After transfecting STAT3 decoy ODN (Dec), scramble ODN (Scr), or Lipofectamine reagent control (Ctrl) into the murine liver cancer cell line Hepa 1–6 for 24 h, miR-146a expression was measured by qPCR analysis. (B–E) Hepa 1–6 cells transfected with miR-146a inhibitors (miR146a-Inh) or control RNA (NC) were injected s.c. into the right posterior flank of C57BL/6 mice (2 × 106 cells/mouse), and tumor-bearing mice were sacrificed after 2 weeks. (B) The inhibitory effect of freshly isolated lymphocytes from liver or spleen on the viability of naïve Hepa 1–6 cells was analyzed by MTT assay at an E:T ratio of 50:1. (C) The weight of tumor, spleen, and liver in the indicated tumor-bearing groups was measured. (D) Flow cytometry analysis was performed to examine CD69, CD25, NKG2D, and FasL levels in CD3−NK1.1+ and CD3+NK1.1− lymphocytes from tumor tissue, spleen, liver, and axillary and inguinal LNs. (E) TGF-β, IL-6, IL-18, TNF-α, and IFN-γ levels in serum were detected by ELISA assay. Data are representative of 3 independent experiments, and statistical significance was determined as **P < 0.01 and *P < 0.05 compared to control.
Similar articles
- miR-451 acts as a suppressor of angiogenesis in hepatocellular carcinoma by targeting the IL-6R-STAT3 pathway.
Liu X, Zhang A, Xiang J, Lv Y, Zhang X. Liu X, et al. Oncol Rep. 2016 Sep;36(3):1385-92. doi: 10.3892/or.2016.4971. Epub 2016 Jul 25. Oncol Rep. 2016. PMID: 27461244 - miR-383 inhibits cell growth and promotes cell apoptosis in hepatocellular carcinoma by targeting IL-17 via STAT3 signaling pathway.
Wang J, Lu L, Luo Z, Li W, Lu Y, Tang Q, Pu J. Wang J, et al. Biomed Pharmacother. 2019 Dec;120:109551. doi: 10.1016/j.biopha.2019.109551. Epub 2019 Oct 21. Biomed Pharmacother. 2019. PMID: 31648164 - LncRNA TUG1 interacting with miR-144 contributes to proliferation, migration and tumorigenesis through activating the JAK2/STAT3 pathway in hepatocellular carcinoma.
Lv J, Kong Y, Gao Z, Liu Y, Zhu P, Yu Z. Lv J, et al. Int J Biochem Cell Biol. 2018 Aug;101:19-28. doi: 10.1016/j.biocel.2018.05.010. Epub 2018 May 20. Int J Biochem Cell Biol. 2018. PMID: 29791864 - Natural bioactive compounds and STAT3 against hepatocellular carcinoma: An update.
Manoharan S, Saha S, Murugesan K, Santhakumar A, Perumal E. Manoharan S, et al. Life Sci. 2024 Jan 15;337:122351. doi: 10.1016/j.lfs.2023.122351. Epub 2023 Dec 15. Life Sci. 2024. PMID: 38103726 Review. - Clinico-Pathological Importance of miR-146a in Lung Cancer.
Wani JA, Majid S, Khan A, Arafah A, Ahmad A, Jan BL, Shah NN, Kazi M, Rehman MU. Wani JA, et al. Diagnostics (Basel). 2021 Feb 10;11(2):274. doi: 10.3390/diagnostics11020274. Diagnostics (Basel). 2021. PMID: 33578944 Free PMC article. Review.
Cited by
- MiRNA-146a/AKT/β-Catenin Activation Regulates Cancer Stem Cell Phenotype in Oral Squamous Cell Carcinoma by Targeting CD24.
Ghuwalewala S, Ghatak D, Das S, Roy S, Das P, Butti R, Gorain M, Nath S, Kundu GC, Roychoudhury S. Ghuwalewala S, et al. Front Oncol. 2021 Oct 12;11:651692. doi: 10.3389/fonc.2021.651692. eCollection 2021. Front Oncol. 2021. PMID: 34712602 Free PMC article. - Essential role of miRNAs in orchestrating the biology of the tumor microenvironment.
Frediani JN, Fabbri M. Frediani JN, et al. Mol Cancer. 2016 May 26;15(1):42. doi: 10.1186/s12943-016-0525-3. Mol Cancer. 2016. PMID: 27231010 Free PMC article. Review. - Tofacitinib Regulates Endostatin via Effects on CD147 and Cathepsin S.
Zisman D, Sabtan H, Rahat MM, Simanovich E, Haddad A, Gazitt T, Feld J, Slobodin G, Kibari A, Elias M, Rahat MA. Zisman D, et al. Int J Mol Sci. 2024 Jul 2;25(13):7267. doi: 10.3390/ijms25137267. Int J Mol Sci. 2024. PMID: 39000375 Free PMC article. - microRNAs Shape Myeloid Cell-Mediated Resistance to Cancer Immunotherapy.
Daveri E, Vergani E, Shahaj E, Bergamaschi L, La Magra S, Dosi M, Castelli C, Rodolfo M, Rivoltini L, Vallacchi V, Huber V. Daveri E, et al. Front Immunol. 2020 Jul 22;11:1214. doi: 10.3389/fimmu.2020.01214. eCollection 2020. Front Immunol. 2020. PMID: 32793185 Free PMC article. Review. - miR-146a Polymorphism (rs2910164) Predicts Colorectal Cancer Patients' Susceptibility to Liver Metastasis.
Iguchi T, Nambara S, Masuda T, Komatsu H, Ueda M, Kidogami S, Ogawa Y, Hu Q, Sato K, Saito T, Hirata H, Sakimura S, Uchi R, Hayashi N, Ito S, Eguchi H, Sugimachi K, Maehara Y, Mimori K. Iguchi T, et al. PLoS One. 2016 Nov 8;11(11):e0165912. doi: 10.1371/journal.pone.0165912. eCollection 2016. PLoS One. 2016. PMID: 27824903 Free PMC article.
References
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA: a cancer journal for clinicians 2010; 60:277-300; PMID:20610543; http://dx.doi.org/10.1017/S000983880999067X - DOI - PubMed
- Lasaro MO, Ertl HC. Targeting inhibitory pathways in cancer immunotherapy. Curr Opin Immunol 2010; 22:385-90; PMID:20466529; http://dx.doi.org/10.1016/j.coi.2010.04.005 - DOI - PMC - PubMed
- Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 2009; 9:798-809; PMID:19851315; http://dx.doi.org/10.1038/nrc2734 - DOI - PMC - PubMed
- Sun X, Sui Q, Zhang C, Tian Z, Zhang J. Targeting blockage of STAT3 in hepatocellular carcinoma cells augments NK cell functions via reverse hepatocellular carcinoma-induced immune suppression. Mol Cancer Ther 2013; 12:2885-96; PMID:24107450; http://dx.doi.org/10.1158/1535-7163.MCT-12-1087 - DOI - PubMed
- Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, Niu G, Kay H, Mule J, Kerr WG, et al. . Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med 2005; 11:1314-21; PMID:16288283; http://dx.doi.org/10.1038/nm1325 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous