Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer - PubMed (original) (raw)
. 2015 Apr 3;348(6230):124-8.
doi: 10.1126/science.aaa1348. Epub 2015 Mar 12.
Matthew D Hellmann 2, Alexandra Snyder 3, Pia Kvistborg 4, Vladimir Makarov 5, Jonathan J Havel 5, William Lee 6, Jianda Yuan 7, Phillip Wong 7, Teresa S Ho 7, Martin L Miller 8, Natasha Rekhtman 9, Andre L Moreira 9, Fawzia Ibrahim 10, Cameron Bruggeman 11, Billel Gasmi 12, Roberta Zappasodi 12, Yuka Maeda 12, Chris Sander 8, Edward B Garon 13, Taha Merghoub 14, Jedd D Wolchok 15, Ton N Schumacher 4, Timothy A Chan 16
Affiliations
- PMID: 25765070
- PMCID: PMC4993154
- DOI: 10.1126/science.aaa1348
Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer
Naiyer A Rizvi et al. Science. 2015.
Abstract
Immune checkpoint inhibitors, which unleash a patient's own T cells to kill tumors, are revolutionizing cancer treatment. To unravel the genomic determinants of response to this therapy, we used whole-exome sequencing of non-small cell lung cancers treated with pembrolizumab, an antibody targeting programmed cell death-1 (PD-1). In two independent cohorts, higher nonsynonymous mutation burden in tumors was associated with improved objective response, durable clinical benefit, and progression-free survival. Efficacy also correlated with the molecular smoking signature, higher neoantigen burden, and DNA repair pathway mutations; each factor was also associated with mutation burden. In one responder, neoantigen-specific CD8+ T cell responses paralleled tumor regression, suggesting that anti-PD-1 therapy enhances neoantigen-specific T cell reactivity. Our results suggest that the genomic landscape of lung cancers shapes response to anti-PD-1 therapy.
Copyright © 2015, American Association for the Advancement of Science.
Figures
Fig. 1. Nonsynonymous mutation burden associated with clinical benefit of anti–PD-1 therapy
(A) Nonsynonymous mutation burden in tumors from patients with DCB (n = 7) or with NDB (n = 9) (median 302 versus 148, Mann-Whitney P = 0.02). (B) PFS in tumors with higher nonsynonymous mutation burden (n = 8) compared to tumors with lower nonsynonymous mutation burden (n = 8) in patients in the discovery cohort (HR 0.19, 95% CI 0.05 to 0.70, log-rank P = 0.01). (C) Nonsynonymous mutation burden in tumors with DCB (n = 7) compared to those with NDB (n = 8) in patients in the validation cohort (median 244 versus 125, Mann-Whitney P = 0.04). (D) PFS in tumors with higher nonsynonymous mutation burden (n = 9) compared to those with lower nonsynonymous mutation burden (n = 9) in patients in the validation cohort (HR 0.15, 95% CI 0.04 to 0.59, log-rank P = 0.006). (E) ROC curve for the correlation of nonsynonymous mutation burden with DCB in discovery cohort. AUC is 0.86 (95% CI 0.66 to 1.05, null hypothesis test P = 0.02). Cut-off of ≥178 nonsynonymous mutations is designated by triangle. (F) Nonsynonymous mutation burden in patients with DCB (n = 14) compared to those with NDB (n = 17) for the entire set of sequenced tumors (median 299 versus 127, Mann-Whitney P = 0.0008). (G) PFS in those with higher nonsynonymous mutation burden (n = 17) compared to those with lower nonsynonymous mutation burden (n = 17) in the entire set of sequenced tumors (HR 0.19, 95% CI 0.08–0.47, log-rank P = 0.0004). In (A), (C), and (F), median and interquartile ranges of total nonsynonymous mutations are shown, with individual values for each tumor shown with dots.
Fig. 2. Molecular smoking signature is significantly associated with improved PFS in NSCLC patients treated with pembrolizumab
PFS in tumors characterized as TH by molecular smoking signature classifier (n = 16) compared to TL tumors (n = 18) (HR 0.15, 95% 0.06 to 0.39, log-rank P = 0.0001).
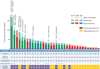
Fig. 3. Mutation burden, clinical response, and factors contributing to mutation burden
Total exonic mutation burden for each sequenced tumor with nonsynonymous (dark shading), synonymous (medium shading), and indels/frameshift mutations (light shading) displayed in the histogram. Columns are shaded to indicate clinical benefit status: DCB, green; NDB, red; not reached 6 months follow-up (NR), blue. The cohort identification (D, discovery; V, validation), best objective response (PR, partial response; SD, stable disease; PD, progression of disease), and PFS (censored at the time of data lock) are reported in the table. Those with ongoing progression-free survival are labeled with ++. The presence of the molecular smoking signature is displayed in the table with TH cases (purple) and TL cases (orange). The presence of deleterious mutations in specific DNA repair/replication genes is indicated by the arrows.
Fig. 4. Candidate neoantigens, neoantigen-specific T cell response, and response to pembrolizumab
(A) Neoantigen burden in patients with DCB (n = 14) compared to NDB (n = 17) across the overall set of sequenced tumors (median 203 versus 83, Mann-Whitney P = 0.001). (B) PFS in tumors with higher candidate neoantigen burden (n = 17) compared to tumors with lower candidate neoantigen burden (n = 17) (HR 0.23, 95%CI 0.09 to 0.58, log-rank P = 0.002). (C) (Top) Representative computed tomography (CT) images of a liver metastasis before and after initiation of treatment. (Middle) Change in radiographic response. (Bottom) Magnitude of the HERC1 P3278S reactive CD8+ T cell response measured in peripheral blood. (D) The proportion of CD8+ T cell population in serially collected autologous PBLs recognizing the HERC1 P3278S neoantigen (ASNA**S**SAAK) before and during pembrolizumab treatment. Each neoantigen is encoded by a unique combination of two fluorescently labeled peptide-MHC complexes (represented individually on each axis); neoantigen-specific T cells are represented by the events in the double positive position indicated with black dots. Percentages indicate the number of CD8+ MHC multimer+ cells out of total CD8 cells. (E) Autologous T cell response to wild-type HERC1 peptide (black), mutant HERC1 P3278S neoantigen (red), or no stimulation (blue), as detected by intracellular cytokine staining. T cell costains for IFNγ and CD8, TNFα, CD107a, and CCL4, respectively, are displayed for the Day 63 and Day 297 time points.
Similar articles
- Late-differentiated effector neoantigen-specific CD8+ T cells are enriched in peripheral blood of non-small cell lung carcinoma patients responding to atezolizumab treatment.
Fehlings M, Jhunjhunwala S, Kowanetz M, O'Gorman WE, Hegde PS, Sumatoh H, Lee BH, Nardin A, Becht E, Flynn S, Ballinger M, Newell EW, Yadav M. Fehlings M, et al. J Immunother Cancer. 2019 Sep 12;7(1):249. doi: 10.1186/s40425-019-0695-9. J Immunother Cancer. 2019. PMID: 31511069 Free PMC article. Clinical Trial. - Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer.
Anagnostou V, Smith KN, Forde PM, Niknafs N, Bhattacharya R, White J, Zhang T, Adleff V, Phallen J, Wali N, Hruban C, Guthrie VB, Rodgers K, Naidoo J, Kang H, Sharfman W, Georgiades C, Verde F, Illei P, Li QK, Gabrielson E, Brock MV, Zahnow CA, Baylin SB, Scharpf RB, Brahmer JR, Karchin R, Pardoll DM, Velculescu VE. Anagnostou V, et al. Cancer Discov. 2017 Mar;7(3):264-276. doi: 10.1158/2159-8290.CD-16-0828. Epub 2016 Dec 28. Cancer Discov. 2017. PMID: 28031159 Free PMC article. - Epigenetic prediction of response to anti-PD-1 treatment in non-small-cell lung cancer: a multicentre, retrospective analysis.
Duruisseaux M, Martínez-Cardús A, Calleja-Cervantes ME, Moran S, Castro de Moura M, Davalos V, Piñeyro D, Sanchez-Cespedes M, Girard N, Brevet M, Giroux-Leprieur E, Dumenil C, Pradotto M, Bironzo P, Capelletto E, Novello S, Cortot A, Copin MC, Karachaliou N, Gonzalez-Cao M, Peralta S, Montuenga LM, Gil-Bazo I, Baraibar I, Lozano MD, Varela M, Ruffinelli JC, Palmero R, Nadal E, Moran T, Perez L, Ramos I, Xiao Q, Fernandez AF, Fraga MF, Gut M, Gut I, Teixidó C, Vilariño N, Prat A, Reguart N, Benito A, Garrido P, Barragan I, Emile JF, Rosell R, Brambilla E, Esteller M. Duruisseaux M, et al. Lancet Respir Med. 2018 Oct;6(10):771-781. doi: 10.1016/S2213-2600(18)30284-4. Epub 2018 Aug 9. Lancet Respir Med. 2018. PMID: 30100403 - PD-1 checkpoint blockade alone or combined PD-1 and CTLA-4 blockade as immunotherapy for lung cancer?
Tanvetyanon T, Gray JE, Antonia SJ. Tanvetyanon T, et al. Expert Opin Biol Ther. 2017 Mar;17(3):305-312. doi: 10.1080/14712598.2017.1280454. Epub 2017 Jan 18. Expert Opin Biol Ther. 2017. PMID: 28064556 Review. - Immunotherapy in non-small cell lung cancer harbouring driver mutations.
Addeo A, Passaro A, Malapelle U, Banna GL, Subbiah V, Friedlaender A. Addeo A, et al. Cancer Treat Rev. 2021 May;96:102179. doi: 10.1016/j.ctrv.2021.102179. Epub 2021 Mar 19. Cancer Treat Rev. 2021. PMID: 33798954 Review. No abstract available.
Cited by
- The oncogenic functions of SPARCL1 in bladder cancer.
Li C, Yuan H, Chen J, Shang K, He H. Li C, et al. J Cell Mol Med. 2024 Nov;28(22):e70196. doi: 10.1111/jcmm.70196. J Cell Mol Med. 2024. PMID: 39548034 Free PMC article. - Machine learning-driven estimation of mutational burden highlights DNAH5 as a prognostic marker in colorectal cancer.
Fang Y, Fu T, Zhang Q, Xiong Z, Yu K, Le A. Fang Y, et al. Biol Direct. 2024 Nov 14;19(1):116. doi: 10.1186/s13062-024-00564-0. Biol Direct. 2024. PMID: 39543663 Free PMC article. - A novel mouse model recapitulating the MMR-defective SCLC subtype uncovers an actionable sensitivity to immune checkpoint blockade.
Ibruli O, Rose F, Beleggia F, Schmitt A, Cartolano M, Fernandez LT, Saggau J, Bonasera D, Kiljan M, Gozum G, Lichius L, Cai J, Niu LN, Caiaffa MI, Herter JM, Walczak H, Liccardi G, Grüll H, Büttner R, Bosco G, George J, Thomas RK, Bozek K, Reinhardt HC, Herter-Sprie GS. Ibruli O, et al. J Cancer Res Clin Oncol. 2024 Nov 14;150(11):496. doi: 10.1007/s00432-024-05942-9. J Cancer Res Clin Oncol. 2024. PMID: 39542886 Free PMC article. - pVACview: an interactive visualization tool for efficient neoantigen prioritization and selection.
Xia H, Hoang MH, Schmidt E, Kiwala S, McMichael J, Skidmore ZL, Fisk B, Song JJ, Hundal J, Mooney T, Walker JR, Goedegebuure SP, Miller CA, Gillanders WE, Griffith OL, Griffith M. Xia H, et al. Genome Med. 2024 Nov 14;16(1):132. doi: 10.1186/s13073-024-01384-7. Genome Med. 2024. PMID: 39538339 Free PMC article. - CAR-engineered NK cells versus CAR T cells in treatment of glioblastoma; strength and flaws.
Sabahi M, Fathi Jouzdani A, Sadeghian Z, Dabbagh Ohadi MA, Sultan H, Salehipour A, Maniakhina L, Rezaei N, Adada B, Mansouri A, Borghei-Razavi H. Sabahi M, et al. J Neurooncol. 2024 Nov 13. doi: 10.1007/s11060-024-04876-z. Online ahead of print. J Neurooncol. 2024. PMID: 39538038 Review.
References
- Coley WB. Clin. Orthop. Relat. Res. 1991;1991(262):3–11. - PubMed
- Robert C, et al. Lancet. 2014;384:1109–1117. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials