GSHSite: exploiting an iteratively statistical method to identify s-glutathionylation sites with substrate specificity - PubMed (original) (raw)
GSHSite: exploiting an iteratively statistical method to identify s-glutathionylation sites with substrate specificity
Yi-Ju Chen et al. PLoS One. 2015.
Abstract
S-glutathionylation, the covalent attachment of a glutathione (GSH) to the sulfur atom of cysteine, is a selective and reversible protein post-translational modification (PTM) that regulates protein activity, localization, and stability. Despite its implication in the regulation of protein functions and cell signaling, the substrate specificity of cysteine S-glutathionylation remains unknown. Based on a total of 1783 experimentally identified S-glutathionylation sites from mouse macrophages, this work presents an informatics investigation on S-glutathionylation sites including structural factors such as the flanking amino acids composition and the accessible surface area (ASA). TwoSampleLogo presents that positively charged amino acids flanking the S-glutathionylated cysteine may influence the formation of S-glutathionylation in closed three-dimensional environment. A statistical method is further applied to iteratively detect the conserved substrate motifs with statistical significance. Support vector machine (SVM) is then applied to generate predictive model considering the substrate motifs. According to five-fold cross-validation, the SVMs trained with substrate motifs could achieve an enhanced sensitivity, specificity, and accuracy, and provides a promising performance in an independent test set. The effectiveness of the proposed method is demonstrated by the correct identification of previously reported S-glutathionylation sites of mouse thioredoxin (TXN) and human protein tyrosine phosphatase 1b (PTP1B). Finally, the constructed models are adopted to implement an effective web-based tool, named GSHSite (http://csb.cse.yzu.edu.tw/GSHSite/), for identifying uncharacterized GSH substrate sites on the protein sequences.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
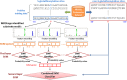
Fig 1. The analytical flowchart of MDDLogo application.
(A) Using chi-square test to detect the maximal dependence of position, and (B) Tree-like visualization of MDDLogo-clustering result.
Fig 2. The conceptual diagram of two-layered SVMs trained with MDDLogo- identified substrate motifs.
Fig 3. TwoSampleLogo presents the compositional biases of amino acids around _S_-glutathionylation sites compared to the non-_S_-glutathionylation sites in mouse macrophages.
The significant amino acids around _S_-glutathionylated cysteine residue is enriched from the positive dataset and presented in upper panel (p < 0.01). Relatively, the high frequency of amino acids around non-_S_-glutathionylated cysteines is depleted from the negative dataset and presented in lower panel.
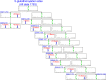
Fig 4. The MDDLogo-clustered subgroups from 1783 _S_-glutathionylation sites in mouse data set.
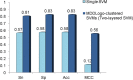
Fig 5. Comparison of independent testing performance between single SVM and MDDLogo-clustered SVM models.
Sn, sensitivity; Sp, specificity; Acc, accuracy; MCC, Matthews Correlation Coefficient.
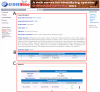
Fig 6. A case study of S-glutathionylation site prediction for mouse thioredoxin (THIO_MOUSE).
The website presents (A) protein information and annotation, (B) S-glutathionylation sites from published experiment, and (C) prediction result of potential consensus motifs.
Similar articles
- SNOSite: exploiting maximal dependence decomposition to identify cysteine S-nitrosylation with substrate site specificity.
Lee TY, Chen YJ, Lu TC, Huang HD, Chen YJ. Lee TY, et al. PLoS One. 2011;6(7):e21849. doi: 10.1371/journal.pone.0021849. Epub 2011 Jul 15. PLoS One. 2011. PMID: 21789187 Free PMC article. - MDD-SOH: exploiting maximal dependence decomposition to identify S-sulfenylation sites with substrate motifs.
Bui VM, Lu CT, Ho TT, Lee TY. Bui VM, et al. Bioinformatics. 2016 Jan 15;32(2):165-72. doi: 10.1093/bioinformatics/btv558. Epub 2015 Sep 26. Bioinformatics. 2016. PMID: 26411868 - dbGSH: a database of S-glutathionylation.
Chen YJ, Lu CT, Lee TY, Chen YJ. Chen YJ, et al. Bioinformatics. 2014 Aug 15;30(16):2386-8. doi: 10.1093/bioinformatics/btu301. Epub 2014 Apr 29. Bioinformatics. 2014. PMID: 24790154 - Glutaredoxin: role in reversible protein s-glutathionylation and regulation of redox signal transduction and protein translocation.
Shelton MD, Chock PB, Mieyal JJ. Shelton MD, et al. Antioxid Redox Signal. 2005 Mar-Apr;7(3-4):348-66. doi: 10.1089/ars.2005.7.348. Antioxid Redox Signal. 2005. PMID: 15706083 Review. - Protein S-glutathionylation: from current basics to targeted modifications.
Popov D. Popov D. Arch Physiol Biochem. 2014 Oct;120(4):123-30. doi: 10.3109/13813455.2014.944544. Epub 2014 Aug 12. Arch Physiol Biochem. 2014. PMID: 25112365 Review.
Cited by
- A novel approach for predicting protein S-glutathionylation.
Anashkina AA, Poluektov YM, Dmitriev VA, Kuznetsov EN, Mitkevich VA, Makarov AA, Petrushanko IY. Anashkina AA, et al. BMC Bioinformatics. 2020 Sep 14;21(Suppl 11):282. doi: 10.1186/s12859-020-03571-w. BMC Bioinformatics. 2020. PMID: 32921310 Free PMC article. - Protein Thiol Redox Signaling in Monocytes and Macrophages.
Short JD, Downs K, Tavakoli S, Asmis R. Short JD, et al. Antioxid Redox Signal. 2016 Nov 20;25(15):816-835. doi: 10.1089/ars.2016.6697. Epub 2016 Jul 13. Antioxid Redox Signal. 2016. PMID: 27288099 Free PMC article. Review. - Protein S-glutathionlyation links energy metabolism to redox signaling in mitochondria.
Mailloux RJ, Treberg JR. Mailloux RJ, et al. Redox Biol. 2016 Aug;8:110-8. doi: 10.1016/j.redox.2015.12.010. Epub 2015 Dec 31. Redox Biol. 2016. PMID: 26773874 Free PMC article. Review. - Proteomic Identification of Protein Glutathionylation in Cardiomyocytes.
VanHecke GC, Abeywardana MY, Ahn YH. VanHecke GC, et al. J Proteome Res. 2019 Apr 5;18(4):1806-1818. doi: 10.1021/acs.jproteome.8b00986. Epub 2019 Mar 11. J Proteome Res. 2019. PMID: 30831029 Free PMC article. - Investigation and identification of protein carbonylation sites based on position-specific amino acid composition and physicochemical features.
Weng SL, Huang KY, Kaunang FJ, Huang CH, Kao HJ, Chang TH, Wang HY, Lu JJ, Lee TY. Weng SL, et al. BMC Bioinformatics. 2017 Mar 14;18(Suppl 3):66. doi: 10.1186/s12859-017-1472-8. BMC Bioinformatics. 2017. PMID: 28361707 Free PMC article.
References
- Dalle-Donne I, Rossi R, Giustarini D, Colombo R, Milzani A (2007) S-glutathionylation in protein redox regulation. Free Radic Biol Med 43: 883–898. - PubMed
- Gallogly MM, Mieyal JJ (2007) Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol 7: 381–391. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous