A conserved function in phosphatidylinositol metabolism for mammalian Vps13 family proteins - PubMed (original) (raw)
A conserved function in phosphatidylinositol metabolism for mammalian Vps13 family proteins
Jae-Sook Park et al. PLoS One. 2015.
Abstract
The Vps13 protein family is highly conserved in eukaryotic cells. In humans, mutations in the gene encoding the family member VPS13A lead to the neurodegenerative disorder chorea-acanthocytosis. In the yeast Saccharomyces cerevisiae, there is just a single version of VPS13, thereby simplifying the task of unraveling its molecular function(s). While VPS13 was originally identified in yeast by its role in vacuolar sorting, recent studies have revealed a completely different function for VPS13 in sporulation, where VPS13 regulates phosphatidylinositol-4-phosphate (PtdIns(4)P) levels in the prospore membrane. This discovery raises the possibility that the disease phenotype associated with vps13A mutants in humans is due to misregulation of PtdIns(4)P in membranes. To determine whether VPS13A affects PtdIns(4)P in membranes from mammalian neuronal cells, phosphatidylinositol phosphate pools were compared in PC12 tissue culture cells in the absence or presence of VPS13A. Consistent with the yeast results, the localization of PtdIns(4)P is specifically altered in VPS13A knockdown cells while other phosphatidylinositol phosphates appear unaffected. In addition, VPS13A is necessary to prevent the premature degeneration of neurites that develop in response to Nerve Growth Factor. The regulation of PtdIns(4)P is therefore a conserved function of the Vps13 family and may play a role in the maintenance of neuronal processes in mammals.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
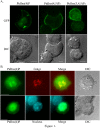
Fig 1. Localization of PtdIns(4)P, PtdIns(4,5)P2 or PtdIns(3,4,5)P3 in PC12 cells.
(A) PC12 cells were transiently transfected with GFP-OSBP-PH, PLCδ1-PH-GFP or Akt-PH-GFP to visualize PtdIns(4)P, PtdIns(4,5)P2 or PtdIns(3,4,5)P3, respectively. (B) PC12 cells were transfected with GFP-OSBP-PH to visualize PtdIns(4)P, fixed and stained with antibodies to the _cis_-Golgi marker GM130 and with DAPI. Representative images are shown. At least 15 cells were scored for each transfection. Scale bars = 5μm.
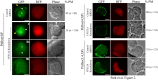
Fig 2. Localization of PtdIns(4)P, PtdIns(4,5)P2 or PtdIns(3,4,5)P3 in PC12 cells carrying scrambled shRNA or VPS13A knockdown shRNA.
PC12 cells were transiently co-transfected with GFP tagged PH domains and vectors expressing either a control, scrambled shRNA or VPS13A shRNA3. Red fluorescence indicates the presence of the shRNA vector. For each sensor, the percentage of knockdown cells in which the sensor was localized at the plasma membrane and the number (n) of cells scored are shown. The data are pooled from three experiments for the PtdIns(4)P and PtdIns(4,5)P2 sensor and two experiments for the PtdIns(3,4,5)P3 sensor. In VPS13A knockdown cells, three patterns of localization for the PtdIns(4)P sensor are seen: class I are similar to the control cells (indicated by “I”); class IIa are lacking in plasma membrane fluorescence but have clear Golgi and nuclear fluorescence (IIa); and class IIb show diffuse fluorescence throughout the cell (IIb). Yellow arrows indicate the plasma membrane. White arrowheads indicate Golgi elements. N = nucleus. Scale bars = 5μm.
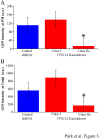
Fig 3. Quantitation of PtdIns(4)P within plasma membranes and the Golgi complex in VPS13A knockdown cells.
(A) The average fluorescence intensity of the GFP-OSBP-PH from the plasma membrane was quantified from cells cotransfected with the control, scrambled shRNA, or the VPS13A knockdown shRNA3. Fluorescence was measured separately for knockdown cells in class I and class IIa (described in legend to Fig 2). Class IIa cells show a significant decrease in plasma membrane fluorescence relative to control (p<0.001, student’s t-test). Error bars indicate standard error. Greater than 12 cells scored in all classes. a.u. = arbitrary units. (B) Average fluorescence intensity of GFP-OSPB-PH from the Golgi complex quantified as in (A). Asterisk indicates significantly reduced fluorescence in the Class IIa cells relative to the control (p<0.01, student’s t-test).
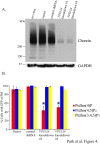
Fig 4. Efficacy of the VPS13A knockdown shRNAs in lentivirus infected cells.
(A) Western blot using anti-chorein antibodies to determine the level chorein expression in control and lentivirus-infected knockdown cells. The levels of GAPDH were used as a loading control for each lysate. For chorein and GAPDH detection, 100μg and 50μg of total lysates were loaded, respectively. (B) Quantitation of PtdIns-phosphate sensor localization to the plasma membrane in in control and lentivirus-infected knockdown cells. Two independent experiments were performed for each reporter. At least 50 cells scored in each experiment. Error bars indicate one standard deviation. Asterisks indicate a significant reduction of PtdIns(4)P sensor localization to the plasma membrane in cells expressing knockdown shRNA #3 or #5 relative to those expressing the scrambled shRNA (p<0.0001, Chi square test).
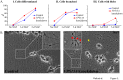
Fig 5. Differentiation and degeneration of VPS13 knockdown PC12 cells upon treatment with NGF.
(A) NGF treated-PC12 cells infected with scrambled or VPS13A shRNAs were fixed on the indicated day and scored for the presence of extended neurites (indicating differentiation, left graph), the fraction of neurites that were branched (middle graph), or the presence of blebs on the neurites. Asterisk indicates a significant difference in the frequency of blebs (p<0.001, Chi square test). The time course was performed twice with similar results, data from one experiment is shown. More than 60 cells were scored for each feature at each time point. (B) Neurite images from lentivirus infected PC12 cells 5 days after NGF treatment. Small boxes are magnified to show the neurites. Red arrows indicate blebs and yellow arrowhead indicates the degenerating neurite in PC12 cells carrying VPS13A shRNA 3Scale bar = 50μm.
Similar articles
- Amino acid substitution equivalent to human chorea-acanthocytosis I2771R in yeast Vps13 protein affects its binding to phosphatidylinositol 3-phosphate.
Rzepnikowska W, Flis K, Kaminska J, Grynberg M, Urbanek A, Ayscough KR, Zoladek T. Rzepnikowska W, et al. Hum Mol Genet. 2017 Apr 15;26(8):1497-1510. doi: 10.1093/hmg/ddx054. Hum Mol Genet. 2017. PMID: 28334785 Free PMC article. - TipC and the chorea-acanthocytosis protein VPS13A regulate autophagy in Dictyostelium and human HeLa cells.
Muñoz-Braceras S, Calvo R, Escalante R. Muñoz-Braceras S, et al. Autophagy. 2015;11(6):918-27. doi: 10.1080/15548627.2015.1034413. Autophagy. 2015. PMID: 25996471 Free PMC article. - Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites.
Park JS, Thorsness MK, Policastro R, McGoldrick LL, Hollingsworth NM, Thorsness PE, Neiman AM. Park JS, et al. Mol Biol Cell. 2016 Aug 1;27(15):2435-49. doi: 10.1091/mbc.E16-02-0112. Epub 2016 Jun 8. Mol Biol Cell. 2016. PMID: 27280386 Free PMC article. - Yeast and other lower eukaryotic organisms for studies of Vps13 proteins in health and disease.
Rzepnikowska W, Flis K, Muñoz-Braceras S, Menezes R, Escalante R, Zoladek T. Rzepnikowska W, et al. Traffic. 2017 Nov;18(11):711-719. doi: 10.1111/tra.12523. Epub 2017 Sep 24. Traffic. 2017. PMID: 28846184 Review. - [Proteins from Vps13 family: from molecular function to pathogenesis of neurodegenerative disorders].
Kamińska J, Kolakowski D. Kamińska J, et al. Postepy Biochem. 2018 Dec 29;64(4):275-287. doi: 10.18388/pb.2018_141. Postepy Biochem. 2018. PMID: 30656912 Review. Polish.
Cited by
- Striking a balance: PIP2 and PIP3 signaling in neuronal health and disease.
Tariq K, Luikart BW. Tariq K, et al. Explor Neuroprotective Ther. 2021;1:86-100. doi: 10.37349/ent.2021.00008. Epub 2021 Oct 29. Explor Neuroprotective Ther. 2021. PMID: 35098253 Free PMC article. - VPS13D promotes peroxisome biogenesis.
Baldwin HA, Wang C, Kanfer G, Shah HV, Velayos-Baeza A, Dulovic-Mahlow M, Brüggemann N, Anding A, Baehrecke EH, Maric D, Prinz WA, Youle RJ. Baldwin HA, et al. J Cell Biol. 2021 May 3;220(5):e202001188. doi: 10.1083/jcb.202001188. J Cell Biol. 2021. PMID: 33891012 Free PMC article. - Yeast-model-based study identified myosin- and calcium-dependent calmodulin signalling as a potential target for drug intervention in chorea-acanthocytosis.
Soczewka P, Kolakowski D, Smaczynska-de Rooij I, Rzepnikowska W, Ayscough KR, Kaminska J, Zoladek T. Soczewka P, et al. Dis Model Mech. 2019 Jan 28;12(1):dmm036830. doi: 10.1242/dmm.036830. Dis Model Mech. 2019. PMID: 30635263 Free PMC article. - Role of VPS13, a protein with similarity to ATG2, in physiology and disease.
Ugur B, Hancock-Cerutti W, Leonzino M, De Camilli P. Ugur B, et al. Curr Opin Genet Dev. 2020 Dec;65:61-68. doi: 10.1016/j.gde.2020.05.027. Epub 2020 Jun 18. Curr Opin Genet Dev. 2020. PMID: 32563856 Free PMC article. Review. - Interaction between VPS13A and the XK scramblase is important for VPS13A function in humans.
Park JS, Hu Y, Hollingsworth NM, Miltenberger-Miltenyi G, Neiman AM. Park JS, et al. J Cell Sci. 2022 Sep 1;135(17):jcs260227. doi: 10.1242/jcs.260227. Epub 2022 Sep 8. J Cell Sci. 2022. PMID: 35950506 Free PMC article.
References
- Velayos-Baeza A, Vettori A, Copley RR, Dobson-Stone C, Monaco AP (2004) Analysis of the human VPS13 gene family. Genomics 84: 536–549. - PubMed
- Nakanishi H, Suda Y, Neiman AM (2007) Erv14 family cargo receptors are necessary for ER exit during sporulation in Saccharomyces cerevisiae . J Cell Sci 120: 908–916. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases