Cdt1-binding protein GRWD1 is a novel histone-binding protein that facilitates MCM loading through its influence on chromatin architecture - PubMed (original) (raw)
. 2015 Jul 13;43(12):5898-911.
doi: 10.1093/nar/gkv509. Epub 2015 May 18.
Kazumitsu Maehara 2, Kazumasa Yoshida 1, Shuhei Yasukouchi 1, Satoko Osano 3, Shinya Watanabe 1, Masahiro Aizawa 1, Takashi Yugawa 3, Tohru Kiyono 3, Hitoshi Kurumizaka 4, Yasuyuki Ohkawa 2, Masatoshi Fujita 5
Affiliations
- PMID: 25990725
- PMCID: PMC4499137
- DOI: 10.1093/nar/gkv509
Cdt1-binding protein GRWD1 is a novel histone-binding protein that facilitates MCM loading through its influence on chromatin architecture
Nozomi Sugimoto et al. Nucleic Acids Res. 2015.
Abstract
Efficient pre-replication complex (pre-RC) formation on chromatin templates is crucial for the maintenance of genome integrity. However, the regulation of chromatin dynamics during this process has remained elusive. We found that a conserved protein, GRWD1 (glutamate-rich WD40 repeat containing 1), binds to two representative replication origins specifically during G1 phase in a CDC6- and Cdt1-dependent manner, and that depletion of GRWD1 reduces loading of MCM but not CDC6 and Cdt1. Furthermore, chromatin immunoprecipitation coupled with high-throughput sequencing (Seq) revealed significant genome-wide co-localization of GRWD1 with CDC6. We found that GRWD1 has histone-binding activity. To investigate the effect of GRWD1 on chromatin architecture, we used formaldehyde-assisted isolation of regulatory elements (FAIRE)-seq or FAIRE-quantitative PCR analyses, and the results suggest that GRWD1 regulates chromatin openness at specific chromatin locations. Taken together, these findings suggest that GRWD1 may be a novel histone-binding protein that regulates chromatin dynamics and MCM loading at replication origins.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Figure 1.
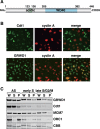
GRWD1 is present in the nucleus throughout interphase and is a chromatin/nuclear matrix-binding protein. (A) Schematic representation of GRWD1 domain structure. An acidic domain and five WD40 repeats are indicated. (B) HeLa cells were double-immunostained with anti-cyclin A and anti-GRWD1 or anti-cyclin A and anti-Cdt1 antibodies. (C) HeLa cells were allowed to grow asynchronously (AS), treated with hydroxyurea (HU) and then released for 2 h (for early S phase) or 9 h (for late S/G2/M phase), and then subjected to chromatin-binding assays. Coomassie Brilliant Blue (CBB) staining served as the loading control. W, whole cell extract; S, Triton X-100-extractable soluble fraction; and P, chromatin/nuclear matrix-bound fraction.
Figure 2.
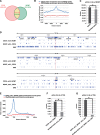
GRWD1 interacts with Cdt1 and CDC6 and binds specifically to replication origins at the lamin B2 and MCM4 loci during G1 phase in a CDC6- and Cdt1-dependent manner. (A) HEK293T cells transfected with empty or T7-Cdt1 expression vectors were immunoprecipitated with anti-GRWD1 antibody or control rabbit IgG. The precipitates (IP) were then immunoblotted with the indicated antibodies. Two percent of the input was also loaded. (B) Nuclear extracts prepared from HEK293T cells were immunoprecipitated with anti-CDC6 antibody. Immunoprecipitates were subjected to immunoblotting with the indicated antibodies. Two percent of the input was also loaded. (C) Soluble extracts from HEK293T cells transfected with the indicated expression vectors were immunoprecipitated with anti-Flag antibody. The first immunoprecipitates were eluted with Flag peptide and then re-immunoprecipitated with anti-T7 antibody. Each precipitate (IP) was then immunoblotted with the indicated antibodies. Fifteen percent of the input for first IP and 10% of the input (i.e. the first precipitates) for second IP were also loaded. (D) GST-Cdt1, GST-CDC6 or GST was incubated with bacterially produced full length GRWD1 (FL) or its truncated mutants (Δacid, lacking the acidic domain; ΔC, lacking C-terminal amino acids 207–446). GST and the associated proteins were then collected on glutathione beads and subjected to immunoblotting with anti-GRWD1 (upper panel) or anti-GST antibody (lower panel). Fifteen percent of the input sample (I) was analyzed with the precipitate (P). (E) Schematic diagram of the origins at the human lamin B2 and MCM4 loci. Positions of qPCR primer pairs used to detect the origins and distal sequences are shown above (black and gray boxes, respectively). Coordinates flanking the origin are given in base pairs. (F) ChIP-qPCR assay with anti-GRWD1 antibody was performed using a previously described set of chromatin samples from asynchronously growing HeLa or T98G cells (15). Results are shown as the percent of input DNA. The means ± SD are shown (HeLa, n = 8; T98G, n = 6). Control IgG gave signals below 0.005% of the inputs (15). (G) ChIP-qPCR assay with anti-GRWD1 antibody was performed using a previously described set of chromatin samples from T98G cells synchronized at the G0, G1/S and late S/G2/M phases (15). The means ± SD are shown (n = 3). (H) ChIP-qPCR assay with anti-GRWD1 antibody was performed using a previously described set of chromatin samples from HeLa cells transfected with control or Cdt1 (siCdt1–1) siRNAs for 48 h (15). Depletion of Cdt1 by specific siRNA, and a corresponding reduction in origin enrichment with anti-Cdt1 and anti-MCM7 antibodies, has already been confirmed (15). The means ± SD are shown (n = 3). (I) ChIP-qPCR assay with anti-GRWD1 antibody was performed for a previously described set of chromatin samples from HEK293T cells transfected with the indicated expression vectors for 48 h (15). The means ± SD are shown (n = 3). (J) HeLa cells transfected with control or CDC6 (mixture of siCDC6–1&3) siRNAs for 48 h were subjected to immunoblotting and ChIP-qPCR assay with the indicated antibodies. For ChIP data, the means ± SD are shown (n = 3).
Figure 3.
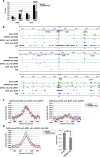
Silencing of GRWD1 suppresses MCM loading and S-phase progression and activates the ATM checkpoint while GRWD1 overexpression alleviates MCM loading defect by Cdt1 inhibition. (A–C) HeLa cells transfected with control or GRWD1 (siGRWD1–3&4) siRNAs for 48 h were subjected to immunoblotting (A), FACS (B) and ChIP-qPCR (C). For ChIP data, the means ± SD are shown (n = 4). (D and E) HeLa cells transfected with the indicated siRNAs for 24 h were arrested in S phase with HU and released. The cells were then harvested at the indicated time points and analyzed by FACS (D) and immunoblotting with indicated antibodies (E). Ser10-phosphorylated-histone H3 was used as a mitotic marker. (F) HFF2/T cells transfected with control (mixture of siLuci&siGFP) or GRWD1 (mixture of siGRWD1–3&4) siRNAs for 48 h were subjected immunoblotting (left panel) or further labeled with 30 μM BrdU for 30 min. The cells were double stained with anti-BrdU antibody and PI and analyzed by FACS (right panels). (G) HeLa cells overexpressing HA-GRWD1 were established by retroviral infection. Cells were then transfected with control (mixture of siLuci&siGFP) or Cdt1 (mixture of siCdt1–4&5) siRNAs for 48 h and then subjected to immunoblotting and ChIP-qPCR assay with the indicated antibodies. For ChIP data, the means ± SD are shown (n = 3).
Figure 4.
GRWD1 Has a Histone-Binding Activity. (A) GST-GRWD1-His, its truncated mutant GST-GRWD1-His Δacid, or GST was mixed with core histones purified from HeLa cells, and proteins bound to glutathione sepharose beads (P) with the input samples (I) were analyzed by CBB staining. (B) GST-GRWD1-His, GST-GRWD1-His Δacid, or GST was mixed with H2A/H2B or H3/H4 complexes, and bound proteins were subjected to CBB staining. (C) Topological assay was carried out with the indicated proteins. Supercoiled DNA (lane 1), DNA relaxed by topoisomerase I (lane 2), relaxed DNA incubated with reaction buffer alone, NAP1 (1 μM), GST (1 μM) or GST-GRWD1-His (0.5 μM or 1 μM), in the presence or absence of core histones, was analyzed on an agarose gel electrophoresis followed by SYBR Gold staining. (D) Mononucleosome assembly assay was performed with the indicated proteins. A 195 bp 5S DNA incubated with reaction buffer alone, NAP1 (1 μM), GST (1 μM) or GST-GRWD1-His (0.5 μM or 1 μM), in the presence or absence of recombinant histones, was analyzed by non-denaturing PAGE.
Figure 5.
Genome wide identification of pre-RC-enriched zones. (A) Venn diagram showing the numbers of CDC6-binding or MCM7-binding peaks (called by MACS2 broad peak caller in ChIP-Seq for asynchronous HeLa cells) and their overlap within 0.5 kb. (B) CDC6 binding peaks were aggregated for MCM7 binding sites (peaks). The average fold-enrichment of CDC6 was plotted relative to the position of all MCM7 binding sites. The ‘shuffled MCM7 peaks’ were randomly chosen from genome. (C) The number of CDC6 peaks within 0.5 kb of MCM7 peaks is significantly higher than that obtained with randomized CDC6 peaks. *, P < 0.001 by Chi-Square test. (D) Distributions of the selected CDC6 (i.e. within 0.5 kb of MCM7) and MCM7 (i.e. within 0.5 kb of CDC6) peaks on genomic loci containing the LMNB2, MCM4 or c-myc genes. Positions of ‘preferential origins’ are indicated. The data for SNS peaks of HeLa cells were derived from a paper by Besnard et al. (39). (E) The frequency of the pre-RC sites (the selected CDC6 peaks that are within 0.5 kb of MCM7 peaks) at all genes was calculated to assess the distribution of the pre-RC sites on genes. Marks -5 kbp and +5 kbp indicate 5 kb upstream of TSSs (Transcription start sites) and 5 kb downstream of TESs (Transcription end sites), respectively. (F) The number of the pre-RC sites (i.e. the selected CDC6 peaks within 0.5 kb of MCM7 peaks) within 0.5 kb of TSS are significantly higher than that obtained with the randomized pre-RC sites. *, P < 0.001 by Chi-Square test. (G) The number of the firing pre-RC sites (i.e. the selected CDC6 peaks which are within 0.5 kb of MCM7 peaks and SNS peaks) within 0.5 kb of TSS are further higher than that obtained with the randomized peaks. *P < 0.001 by Chi-Square test.
Figure 6.
GRWD1 co-localizes with pre-RC genome wide. (A) Visual representation of the GRWD1-binding sites (the selected GRWD1 [sGRWD1] peaks, which are within 0.5 kb of HA-GRWD1 peaks) from asynchronous HeLa cells for genomic loci containing the LMNB2, MCM4 or c-myc genes. For comparison, the data shown in Figure 5D are re-presented in parallel. (B) Venn diagram showing the numbers of the pre-RC sites (CDC6_w0.5_MCM7) or the selected GRWD1-binding sites and their overlap within 0.5 kb. (C) The selected GRWD1 binding sites (peaks) were aggregated for the pre-RC sites (CDC6_w0.5_MCM7). The average fold-enrichment of GRWD1 was plotted relative to the position of the pre-RC sites. (D) The number of the pre-RC sites within 0.5 kb of the selected GRWD1 peaks is significantly higher than that obtained with the randomized pre-RC sites. *, P < 0.001 by Chi-Square test. (E) The number of the firing pre-RC sites (CDC6 peaks that are within 0.5 kb of MCM7 and SNS) within 0.5 kb of the selected GRWD1 peaks is significantly higher than that obtained with the randomized firing pre-RC sites. *, P < 0.001 by Chi-Square test. (F) The pre-RC sites with GRWD1 (CDC6_w0.5_MCM7_w0.5_sGRWD1) appear to be more closely associated with SNS than the pre-RC sites without GRWD1.
Figure 7.
GRWD1 affects chromatin architecture (chromatin openness) at pre-RC sites. (A) GRWD1 depletion reduces chromatin openness at the replication origins at the lamin B2 and MCM4 loci. Asynchronous HeLa cells were treated with control or GRWD1 (mixture of siGRWD1–3&4) siRNAs for 48 h and subjected to FAIRE-qPCR. The means ± SD are shown (n = 3). (B) Visual representation of FAIRE-Seq results for genomic loci containing the LMNB2, MCM4 or c-myc genes. The FAIRE DNA from HeLa cells treated with control (mixture of siLuci&siGFP) or GRWD1 (mixture of siGRWD1–3&4) siRNAs for 48 h was subjected to Seq. For comparison, the data shown in Figures 5D and 6A are re-presented in parallel. *Examples of the pre-RC sites with GRWD1 where FAIRE profiles appear to be affected by GRWD1 depletion. **Examples of the pre-RC sites with GRWD1 where FAIRE profiles appear not to be affected by GRWD1 depletion. (C) Cumulative FAIRE signal profiles at the pre-RC sites with GRWD1 (CDC6_w0.5_MCM7_w0.5_sGRWD1) (left) and those without GRWD1 (right) are shown for either control or GRWD1 siRNA-treated HeLa cells. For both sites, 10 000 sites were randomly selected from the corresponding data sets and used to adjust the peak numbers for the aggregation plots. (D) GRWD1 depletion significantly affects FAIRE profiles around the firing pre-RC sites with GRWD1. Cumulative FAIRE signal profiles at the firing pre-RC sites with GRWD1 (CDC6_w0.5_MCM7_w0.5_sGRWD1_w0.5_SNS; 10 631 peaks) are shown for either control or GRWD1 siRNA-treated HeLa cells (left). Average FAIRE signal of the 10 631 peaks (± 2 kb from the center of each peaks) from GRWD1-depleted cells is significantly lower than that from control siRNA-treated cells (right panel). *, P < 0.001.
Similar articles
- Nucleosome assembly and disassembly activity of GRWD1, a novel Cdt1-binding protein that promotes pre-replication complex formation.
Aizawa M, Sugimoto N, Watanabe S, Yoshida K, Fujita M. Aizawa M, et al. Biochim Biophys Acta. 2016 Nov;1863(11):2739-2748. doi: 10.1016/j.bbamcr.2016.08.008. Epub 2016 Aug 20. Biochim Biophys Acta. 2016. PMID: 27552915 - Molecular Mechanism for Chromatin Regulation During MCM Loading in Mammalian Cells.
Sugimoto N, Fujita M. Sugimoto N, et al. Adv Exp Med Biol. 2017;1042:61-78. doi: 10.1007/978-981-10-6955-0_3. Adv Exp Med Biol. 2017. PMID: 29357053 Review. - H3K9me3 demethylase Kdm4d facilitates the formation of pre-initiative complex and regulates DNA replication.
Wu R, Wang Z, Zhang H, Gan H, Zhang Z. Wu R, et al. Nucleic Acids Res. 2017 Jan 9;45(1):169-180. doi: 10.1093/nar/gkw848. Epub 2016 Sep 26. Nucleic Acids Res. 2017. PMID: 27679476 Free PMC article. - Identification and characterization of a novel component of the human minichromosome maintenance complex.
Sakwe AM, Nguyen T, Athanasopoulos V, Shire K, Frappier L. Sakwe AM, et al. Mol Cell Biol. 2007 Apr;27(8):3044-55. doi: 10.1128/MCB.02384-06. Epub 2007 Feb 12. Mol Cell Biol. 2007. PMID: 17296731 Free PMC article. - Cdt1 and geminin in DNA replication initiation.
Caillat C, Perrakis A. Caillat C, et al. Subcell Biochem. 2012;62:71-87. doi: 10.1007/978-94-007-4572-8_5. Subcell Biochem. 2012. PMID: 22918581 Review.
Cited by
- Quantity and quality of minichromosome maintenance protein complexes couple replication licensing to genome integrity.
Yadav AK, Polasek-Sedlackova H. Yadav AK, et al. Commun Biol. 2024 Feb 9;7(1):167. doi: 10.1038/s42003-024-05855-w. Commun Biol. 2024. PMID: 38336851 Free PMC article. Review. - GRWD1 Over-Expression Promotes Gastric Cancer Progression by Activating Notch Signaling Pathway via Up-Regulation of ADAM17.
Ding H, Feng Z, Hu K. Ding H, et al. Dig Dis Sci. 2024 Mar;69(3):821-834. doi: 10.1007/s10620-023-08208-5. Epub 2024 Jan 3. Dig Dis Sci. 2024. PMID: 38172445 - DNA replication and replication stress response in the context of nuclear architecture.
González-Acosta D, Lopes M. González-Acosta D, et al. Chromosoma. 2024 Jan;133(1):57-75. doi: 10.1007/s00412-023-00813-7. Epub 2023 Dec 6. Chromosoma. 2024. PMID: 38055079 Free PMC article. Review. - Landscape of RNA-binding proteins in diagnostic utility, immune cell infiltration and PANoptosis features of heart failure.
Li J, Zhang X, Ren P, Wu Y, Wang Y, Zhou W, Wang Z, Chao P. Li J, et al. Front Genet. 2022 Oct 14;13:1004163. doi: 10.3389/fgene.2022.1004163. eCollection 2022. Front Genet. 2022. PMID: 36313471 Free PMC article. - Super-Enhancer-Associated nine-gene prognostic score model for prediction of survival in chronic lymphocytic leukemia patients.
Liang X, Meng Y, Li C, Liu L, Wang Y, Pu L, Hu L, Li Q, Zhai Z. Liang X, et al. Front Genet. 2022 Sep 15;13:1001364. doi: 10.3389/fgene.2022.1001364. eCollection 2022. Front Genet. 2022. PMID: 36186463 Free PMC article.
References
- Arias E.E., Walter J.C. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. - PubMed
- Diffley J.F. Regulation of early events in chromosome replication. Curr. Biol. 2004;14:R778–R786. - PubMed
- Masai H., Matsumoto S., You Z., Yoshizawa-Sugata N., Oda M. Eukaryotic chromosome DNA replication: Where, when, and how. Annu. Rev. Biochem. 2010;79:89–130. - PubMed
- Sugimoto N., Yoshida K., Tatsumi Y., Yugawa T., Narisawa-Saito M., Waga S., Kiyono T., Fujita M. Redundant and differential regulation of multiple licensing factors ensures prevention of re-replication in normal human cells. J. Cell. Sci. 2009;122:1184–1191. - PubMed
- Vaziri C., Saxena S., Jeon Y., Lee C., Murata K., Machida Y., Wagle N., Hwang D.S., Dutta A. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell. 2003;11:997–1008. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases