Comparative Analysis of Anther Transcriptome Profiles of Two Different Rice Male Sterile Lines Genotypes under Cold Stress - PubMed (original) (raw)
Comparative Analysis of Anther Transcriptome Profiles of Two Different Rice Male Sterile Lines Genotypes under Cold Stress
Bin Bai et al. Int J Mol Sci. 2015.
Abstract
Rice is highly sensitive to cold stress during reproductive developmental stages, and little is known about the mechanisms of cold responses in rice anther. Using the HiSeq™ 2000 sequencing platform, the anther transcriptome of photo thermo sensitive genic male sterile lines (PTGMS) rice Y58S and P64S (Pei'ai64S) were analyzed at the fertility sensitive stage under cold stress. Approximately 243 million clean reads were obtained from four libraries and aligned against the oryza indica genome and 1497 and 5652 differentially expressed genes (DEGs) were identified in P64S and Y58S, respectively. Both gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were conducted for these DEGs. Functional classification of DEGs was also carried out. The DEGs common to both genotypes were mainly involved in signal transduction, metabolism, transport, and transcriptional regulation. Most of the DEGs were unique for each comparison group. We observed that there were more differentially expressed MYB (Myeloblastosis) and zinc finger family transcription factors and signal transduction components such as calmodulin/calcium dependent protein kinases in the Y58S comparison group. It was also found that ribosome-related DEGs may play key roles in cold stress signal transduction. These results presented here would be particularly useful for further studies on investigating the molecular mechanisms of rice responses to cold stress.
Keywords: PTGMS rice; RNA-Seq; anther; cold stress; transcriptome.
Figures
Figure 1
The comparison of the seed setting rates of Y58S and P64S in the control and in the cold treatment. CK indicates control treatment; CT indicates cold treatment. Vertical bar indicates standard error.
Figure 2
Hierarchical cluster analysis of gene expression based on the log ratio fold change data. The color scale indicates the gene expression level: red color represents increased transcript abundance and blue color represents decreased transcript abundance. TP and CP indicate cold treated P64S and control treated P64S, respectively. TY and CY indicate cold treated Y58S and control treated Y58S, respectively.
Figure 3
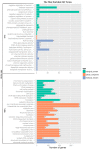
Gene ontology (GO) classifications of differentially expressed genes (DEGs) in P64S and Y58S, respectively. * indicates significantly enriched GO terms with correct _p_-value < 0.05.
Figure 4
Comparison of genes expression levels using RNA-Seq and qRT-PCR. The relative expression values were normalized to the rice actin 1 gene. Error bars indicate standard deviation. Log2FC refers to the log2 fold change.
Similar articles
- Transcriptomics profiling in response to cold stress in cultivated rice and weedy rice.
Guan S, Xu Q, Ma D, Zhang W, Xu Z, Zhao M, Guo Z. Guan S, et al. Gene. 2019 Feb 15;685:96-105. doi: 10.1016/j.gene.2018.10.066. Epub 2018 Oct 30. Gene. 2019. PMID: 30389557 - Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis.
Pradhan SK, Pandit E, Nayak DK, Behera L, Mohapatra T. Pradhan SK, et al. BMC Plant Biol. 2019 Aug 14;19(1):352. doi: 10.1186/s12870-019-1922-8. BMC Plant Biol. 2019. PMID: 31412781 Free PMC article. - Integrated RNA-Seq Analysis and Meta-QTLs Mapping Provide Insights into Cold Stress Response in Rice Seedling Roots.
Kong W, Zhang C, Qiang Y, Zhong H, Zhao G, Li Y. Kong W, et al. Int J Mol Sci. 2020 Jun 29;21(13):4615. doi: 10.3390/ijms21134615. Int J Mol Sci. 2020. PMID: 32610550 Free PMC article. - Genome-wide transcriptome profiling provides insights into panicle development of rice (Oryza sativa L.).
Ke S, Liu XJ, Luan X, Yang W, Zhu H, Liu G, Zhang G, Wang S. Ke S, et al. Gene. 2018 Oct 30;675:285-300. doi: 10.1016/j.gene.2018.06.105. Epub 2018 Jun 30. Gene. 2018. PMID: 29969697 - Genome-wide transcriptome analysis of female-sterile rice ovule shed light on its abortive mechanism.
Yang L, Wu Y, Yu M, Mao B, Zhao B, Wang J. Yang L, et al. Planta. 2016 Nov;244(5):1011-1028. doi: 10.1007/s00425-016-2563-x. Epub 2016 Jun 29. Planta. 2016. PMID: 27357232
Cited by
- Comparative transcriptome analysis and meta-QTLs mapping reveal the regulatory mechanism of cold tolerance in rice at the budding stage.
Li N, Miao J, Li Y, Ji F, Yang M, Dai K, Zhou Z, Hu D, Guo H, Fang H, Wang H, Wang M, Yang J. Li N, et al. Heliyon. 2024 Sep 14;10(18):e37933. doi: 10.1016/j.heliyon.2024.e37933. eCollection 2024 Sep 30. Heliyon. 2024. PMID: 39328527 Free PMC article. - AP2/EREBP Pathway Plays an Important Role in Chaling Wild Rice Tolerance to Cold Stress.
Yang S, Zhou J, Li Y, Wu J, Ma C, Chen Y, Sun X, Wu L, Liang X, Fu Q, Xu Z, Li L, Huang Z, Zhu J, Jia X, Ye X, Chen R. Yang S, et al. Int J Mol Sci. 2023 Sep 22;24(19):14441. doi: 10.3390/ijms241914441. Int J Mol Sci. 2023. PMID: 37833888 Free PMC article. - Cold-responsive transcription factors in Arabidopsis and rice: A regulatory network analysis using array data and gene co-expression network.
Edrisi Maryan K, Farrokhi N, Samizadeh Lahiji H. Edrisi Maryan K, et al. PLoS One. 2023 Jun 8;18(6):e0286324. doi: 10.1371/journal.pone.0286324. eCollection 2023. PLoS One. 2023. PMID: 37289769 Free PMC article. - Oligomeric Proanthocyanidins Confer Cold Tolerance in Rice through Maintaining Energy Homeostasis.
Li J, Feng B, Yu P, Fu W, Wang W, Lin J, Qin Y, Li H, Chen T, Xu C, Tao L, Wu Z, Fu G. Li J, et al. Antioxidants (Basel). 2022 Dec 29;12(1):79. doi: 10.3390/antiox12010079. Antioxidants (Basel). 2022. PMID: 36670941 Free PMC article. - Isolation and characterization of wheat ice recrystallisation inhibition gene promoter involved in low temperature and methyl jasmonate responses.
Jin Y, Ding X, Li J, Guo Z. Jin Y, et al. Physiol Mol Biol Plants. 2022 Dec;28(11-12):1969-1979. doi: 10.1007/s12298-022-01257-6. Epub 2022 Dec 6. Physiol Mol Biol Plants. 2022. PMID: 36573144 Free PMC article.
References
- Ito Y., Katsura K., Maruyama K., Taji T., Kobayashi M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006;47:141–153. doi: 10.1093/pcp/pci230. - DOI - PubMed
- Maruyama K., Todaka D., Mizoi J., Yoshida T., Kidokoro S., Matsukura S., Takasaki H., Sakurai T., Yamamoto Y.Y., Yoshiwara K., et al. Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 2012;19:37–49. doi: 10.1093/dnares/dsr040. - DOI - PMC - PubMed
- Rabbani M.A., Maruyama K., Abe H., Khan M.A., Katsura K., Ito Y., Yoshiwara K., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003;133:1755–1767. doi: 10.1104/pp.103.025742. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources