TipC and the chorea-acanthocytosis protein VPS13A regulate autophagy in Dictyostelium and human HeLa cells - PubMed (original) (raw)
TipC and the chorea-acanthocytosis protein VPS13A regulate autophagy in Dictyostelium and human HeLa cells
Sandra Muñoz-Braceras et al. Autophagy. 2015.
Abstract
Deficient autophagy causes a distinct phenotype in Dictyostelium discoideum, characterized by the formation of multitips at the mound stage. This led us to analyze autophagy in a number of multitipped mutants described previously (tipA(-), tipB(-), tipC(-), and tipD(-)). We found a clear autophagic dysfunction in tipC(-) and tipD(-) while the others showed no defects. tipD codes for a homolog of Atg16, which confirms the role of this protein in Dictyostelium autophagy and validates our approach. The tipC-encoded protein is highly similar to human VPS13A (also known as chorein), whose mutations cause the chorea-acanthocytosis syndrome. No member of the VPS13 protein family has been previously related to autophagy despite the presence of a region of similarity to Atg2 at the C terminus. This region also contains the conserved domain of unknown function DUF1162. Of interest, the expression of the TipC C-terminal coding sequence containing these 2 motifs largely complemented the mutant phenotype. Dictyostelium cells lacking TipC displayed a reduced number of autophagosomes visualized with the markers GFP-Atg18 and GFP-Atg8 and an impaired autophagic degradation as determined by a proteolytic cleavage assay. Downregulation of human VPS13A in HeLa cells by RNA interference confirmed the participation of the human protein in autophagy. VPS13A-depleted cells showed accumulation of autophagic markers and impaired autophagic flux.
Keywords: ATG, autophagy related; AX4, axenic strain 4; DUF, domain of unknown function; Dictyostelium; GFP, green fluorescent protein; HeLa cells; LC3, microtubule-associated protein 1 light chain 3; PtdIns3K, phosphatidylinositol 3-kinase; PtdIns3P, phosphatidylinositol 3-phosphate; TipC; VPS13; VPS13, vacuolar protein sorting 13 homolog (S. cerevisiae); VPS13A; WIPI, WD repeat domain, phosphoinoside interacting; autophagy; chorea-acanthocytosis; chorein.
Figures
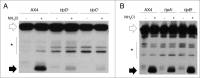
Figure 1.
Autophagic flux is decreased in _tipC_− and _tipD_− cells. Proteolytic cleavage assay was performed in strains transfected with the marker GFP-Tkt-1. Protein extracts were analyzed by western blot using anti-GFP antibody. (A) The accumulation of cleaved GFP fragment (black arrows) in the presence of 100 mM NH4Cl is reduced in _tipC_− and _tipD_− mutants compared to the wild-type AX4 strain. (B) _tipA_− and _tipB_− levels of cleaved GFP are similar to those of AX4. The complete GFP-Tkt-1fusion protein is marked by white arrows and the stars indicate nonspecific immunoreactive bands.
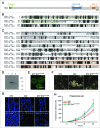
Figure 2 (See previous page).
The conserved C terminus of TipC largely complements the _tipC_− phenotype. (A) Scheme of the 3848-amino acid TipC protein and the conserved domains (colored boxes). The line under the scheme depicts the fragment expressed in the complementation experiments. Alignment of the N-terminal (B) and C-terminal (C) regions of D. discoideum TipC (EAL73163.1) and the human VPS13A (NP_150648.2) proteins using the ClustalW algorithm and shaded using the Boxshade tool at the SDSC Biology WorkBench server (
). Identical residues are shaded black, and similar residues are shaded gray. Colored boxes frame the conserved domains. (D) Western blot using anti-GFP antibody confirmed the presence of a unique band of 150 kDa in the transformed strain with the C-terminal region of TipC (amino acids 2725 to 3848) fused to GFP. Lanes 1 and 3 correspond to protein extracts of AX4 and _tipC_− that do not express the fused proteins and were used as controls of antibody specificity. (E) The C-terminal TipC polypeptide fused to GFP is localized in the cytoplasm according to in vivo confocal microscopy visualization of transformed cells. (F) The complemented strain rescues fruiting body development. Scale bar: 10 μm. (G) Calcofluor staining shows details of stalk differentiation and spore shape of the different strains. For _tipC_− mainly cell debris could be visualized. Scale bar: 10 μm. Spore efficiency of each strain (sp. eff.) is indicated. (H) Phagocytosis rate of the wild-type, the mutant, and the complemented strain was measured through the internalized fluorescent signal of cells that were previously incubated with fluorescent beads during the indicated times.
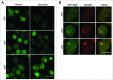
Figure 3.
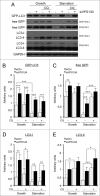
The pattern of GFP-Atg8 is altered in _tipC_− and _tipD_− cells. (A) In vivo confocal analysis of cells expressing GFP-Atg8 in growing and starvation conditions. Puncta formation is inhibited in starved _tipC_− cells compared with AX4. In contrast, large aggregates of the autophagosome marker are evident in _tipD_− cells in both growing conditions. (B) Immunofluorescence of ubiquitin in the cells expressing GFP-Atg8 demonstrates the presence of ubiquitin-positive protein aggregates in both mutants. The ubiquitin structures are smaller in _tipC_− cells and do not always colocalize with GFP-Atg8, while the autophagosome marker is clearly contained in the large _tipD_− ubiquitinated aggregates. Scale bar: 10 μm.
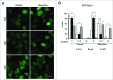
Figure 4.
GFP-Atg18 puncta signaling is decreased in _tipC_− cells and accumulates in _tipD_− cells. (A) Cells expressing GFP-Atg18 were analyzed in vivo by confocal microscopy. Puncta formation is induced under starvation conditions in AX4 cells. However, most of _tipC_− cells do not display any puncta. Conversely, many puncta are observed _tipD_− cells regardless of the nutritional conditions. Scale bar: 10 μm. (B) Quantification of GFP-Atg18 puncta of at least 100 cells in blinded images from 3 independent experiments. Results are shown as mean values with standard deviations of the percentage of cells showing the indicated number of puncta. Asterisks indicate the level of significance of the Student t test (* < 0.05; ** < 0.01).
Figure 5.
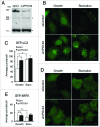
VPS13A downregulation causes GFP-LC3 and GFP-WIPI1 accumulation. (A) Western blot of control and VPS13A siRNA-transfected cells. A band of the VPS13A-predicted molecular weight decreases in the _VPS13A_-silenced cells. The asterisks indicate nonspecific immunoreactive bands. (B and C) HeLa cells stably expressing GFP-LC3 were transfected with control or VPS13A siRNAs and the pattern observed by confocal microscopy. _VPS13A_-depleted cells show accumulated puncta and less translocation from the nucleus to the cytoplasm during starvation. (D and E) HeLa cells were transfected with GFP-WIPI1 and the pattern observed by confocal microscopy. The number of puncta is higher in VPS13A siRNA-treated cells and does not significantly increase following starvation. The graphs show the mean and the standard deviation of puncta quantification of more than 40 cells from 3 independent experiments. Asterisks indicate the level of significance of the Student t test (* < 0.05; ** < 0.01). Scale bar: 10 μm.
Figure 6.
A reduced autophagic flux is observed in VPS13A siRNA-treated cells. (A) GFP-LC3, free GFP, and endogenous LC3-I/-II were analyzed by western blot from control or VPS13A siRNA-treated cells under growth or starvation conditions with or without 5 μM chloroquine (CQ). (B–E) Densitometric quantification showing the mean and the standard deviations of at least 3 independent experiments. The data was normalized against GAPDH. Asterisks indicate the level of significance of the Student t test (* < 0.05; ** < 0.01; *** < 0.001).
Similar articles
- Yeast and other lower eukaryotic organisms for studies of Vps13 proteins in health and disease.
Rzepnikowska W, Flis K, Muñoz-Braceras S, Menezes R, Escalante R, Zoladek T. Rzepnikowska W, et al. Traffic. 2017 Nov;18(11):711-719. doi: 10.1111/tra.12523. Epub 2017 Sep 24. Traffic. 2017. PMID: 28846184 Review. - VPS13A is closely associated with mitochondria and is required for efficient lysosomal degradation.
Muñoz-Braceras S, Tornero-Écija AR, Vincent O, Escalante R. Muñoz-Braceras S, et al. Dis Model Mech. 2019 Feb 22;12(2):dmm036681. doi: 10.1242/dmm.036681. Dis Model Mech. 2019. PMID: 30709847 Free PMC article. - tip genes act in parallel pathways of early Dictyostelium development.
Stege JT, Laub MT, Loomis WF. Stege JT, et al. Dev Genet. 1999;25(1):64-77. doi: 10.1002/(SICI)1520-6408(1999)25:1<64::AID-DVG7>3.0.CO;2-1. Dev Genet. 1999. PMID: 10402673 - The phenotypes of ATG9, ATG16 and ATG9/16 knock-out mutants imply autophagy-dependent and -independent functions.
Xiong Q, Ünal C, Matthias J, Steinert M, Eichinger L. Xiong Q, et al. Open Biol. 2015 Apr;5(4):150008. doi: 10.1098/rsob.150008. Open Biol. 2015. PMID: 25878144 Free PMC article. - Physiological and Pathogenesis Significance of Chorein in Health and Disease.
Alkahtani S, Alkahtane AA, Alarifi S. Alkahtani S, et al. Physiol Res. 2024 Apr 30;73(2):189-203. doi: 10.33549/physiolres.935268. Physiol Res. 2024. PMID: 38710051 Free PMC article. Review.
Cited by
- Dissecting the function of Atg1 complex in Dictyostelium autophagy reveals a connection with the pentose phosphate pathway enzyme transketolase.
Mesquita A, Tábara LC, Martinez-Costa O, Santos-Rodrigo N, Vincent O, Escalante R. Mesquita A, et al. Open Biol. 2015 Aug;5(8):150088. doi: 10.1098/rsob.150088. Open Biol. 2015. PMID: 26246495 Free PMC article. - Yeast as a Model to Find New Drugs and Drug Targets for _VPS13_-Dependent Neurodegenerative Diseases.
Kaminska J, Soczewka P, Rzepnikowska W, Zoladek T. Kaminska J, et al. Int J Mol Sci. 2022 May 4;23(9):5106. doi: 10.3390/ijms23095106. Int J Mol Sci. 2022. PMID: 35563497 Free PMC article. Review. - The WIPI Gene Family and Neurodegenerative Diseases: Insights From Yeast and Dictyostelium Models.
Vincent O, Antón-Esteban L, Bueno-Arribas M, Tornero-Écija A, Navas MÁ, Escalante R. Vincent O, et al. Front Cell Dev Biol. 2021 Sep 1;9:737071. doi: 10.3389/fcell.2021.737071. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34540850 Free PMC article. Review. - Drosophila Vps13 Is Required for Protein Homeostasis in the Brain.
Vonk JJ, Yeshaw WM, Pinto F, Faber AI, Lahaye LL, Kanon B, van der Zwaag M, Velayos-Baeza A, Freire R, van IJzendoorn SC, Grzeschik NA, Sibon OC. Vonk JJ, et al. PLoS One. 2017 Jan 20;12(1):e0170106. doi: 10.1371/journal.pone.0170106. eCollection 2017. PLoS One. 2017. PMID: 28107480 Free PMC article. - Ultrastructural Abnormalities in Induced Pluripotent Stem Cell-Derived Neural Stem Cells and Neurons of Two Cohen Syndrome Patients.
Shnaider TA, Khabarova AA, Morozova KN, Yunusova AM, Yakovleva SA, Chvileva AS, Wolf ER, Kiseleva EV, Grigor'eva EV, Voinova VY, Lagarkova MA, Pomerantseva EA, Musatova EV, Smirnov AV, Smirnova AV, Stoklitskaya DS, Arefieva TI, Larina DA, Nikitina TV, Pristyazhnyuk IE. Shnaider TA, et al. Cells. 2023 Nov 25;12(23):2702. doi: 10.3390/cells12232702. Cells. 2023. PMID: 38067130 Free PMC article.
References
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 2010; 22: 124-31; PMID:20034776; http://dx.doi.org/10.1016/j.ceb.2009.11.014 - DOI - PMC - PubMed
- Calvo-Garrido J, Carilla-Latorre S, Kubohara Y, Santos-Rodrigo N, Mesquita A, Soldati T, Golstein P, Escalante R. Autophagy in Dictyostelium: genes and pathways, cell death and infection. Autophagy 2010; 6: 686-701; PMID:20603609; http://dx.doi.org/10.4161/auto.6.6.12513 - DOI - PubMed
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107-32; PMID:21801009; http://dx.doi.org/10.1146/annurev-cellbio-092910-154005 - DOI - PubMed
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182:685-701; PMID:18725538; http://dx.doi.org/10.1083/jcb.200803137 - DOI - PMC - PubMed
- Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010; 6:764-76; PMID:20639694; http://dx.doi.org/10.4161/auto.6.6.12709 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials