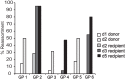Influenza A Virus Coinfection through Transmission Can Support High Levels of Reassortment - PubMed (original) (raw)
Influenza A Virus Coinfection through Transmission Can Support High Levels of Reassortment
Hui Tao et al. J Virol. 2015 Aug.
Abstract
The reassortment of gene segments between influenza viruses increases genomic diversity and plays an important role in viral evolution. We have shown previously that this process is highly efficient within a coinfected cell and, given synchronous coinfection at moderate or high doses, can give rise to ~60 to 70% of progeny shed from an animal host. Conversely, reassortment in vivo can be rendered undetectable by lowering viral doses or extending the time between infections. One might also predict that seeding of transmitted viruses into different sites within the target tissue could limit subsequent reassortment. Given the potential for stochastic factors to restrict reassortment during natural infection, we sought to determine its efficiency in a host coinfected through transmission. Two scenarios were tested in a guinea pig model, using influenza A/Panama/2007/99 (H3N2) virus (wt) and a silently mutated variant (var) thereof as parental virus strains. In the first, coinfection was achieved by exposing a naive guinea pig to two cagemates, one infected with wt and the other with var virus. When such exposure led to coinfection, robust reassortment was typically seen, with 50 to 100% of isolates carrying reassortant genomes at one or more time points. In the second scenario, naive guinea pigs were exposed to a cagemate that had been coinoculated with wt and var viruses. Here, reassortment occurred in the coinoculated donor host, multiple variants were transmitted, and reassortants were prevalent in the recipient host. Together, these results demonstrate the immense potential for reassortment to generate viral diversity in nature.
Importance: Influenza viruses evolve rapidly under selection due to the generation of viral diversity through two mechanisms. The first is the introduction of random errors into the genome by the viral polymerase, which occurs with a frequency of approximately 10(-5) errors/nucleotide replicated. The second is reassortment, or the exchange of gene segments between viruses. Reassortment is known to occur readily under well-controlled laboratory conditions, but its frequency in nature is not clear. Here, we tested the hypothesis that reassortment efficiency following coinfection through transmission would be reduced compared to that seen with coinoculation. Contrary to this hypothesis, our results indicate that coinfection achieved through transmission supports high levels of reassortment. These results suggest that reassortment is not exquisitely sensitive to stochastic effects associated with transmission and likely occurs in nature whenever a host is infected productively with more than one influenza A virus.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
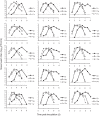
FIG 1
Nasal-wash titers of donor and recipient guinea pigs in a dual-exposure model. Each cage of three guinea pigs is represented by a separate graph. The symbols in each graph indicate the identification (ID) number of each animal, which comprises the cage number followed by “r” for a recipient guinea pig, “w” for a wt-virus-infected donor, or “v” for a var-virus-infected donor. Transmission in cages 1 to 6 was performed in parallel and involved Pan/99wt and Pan/99var6 viruses. Transmission in cages 7 to 15 was performed in parallel at a later time and involved Pan/99wt and Pan/99var15 viruses. The titers of recipient guinea pigs are plotted with solid lines and circles. Donor guinea pigs are shown with open symbols and dashed lines. The limit of detection (50 PFU/ml) is indicated with a horizontal dashed line, and titers below the limit of detection were plotted as 45 PFU/ml. d, day(s).
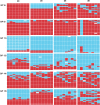
FIG 2
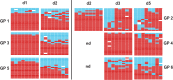
Viral genotypes sampled from nasal washes of guinea pigs coinfected through two independent transmission events. Genotype tables are shown for six guinea pigs coinfected with wt and var viruses through contact exposure to singly inoculated donor hosts. The day postinoculation on which each nasal wash was collected is indicated at the top and refers to the day after inoculation of the donor guinea pigs. The guinea pig (GP) ID numbers are shown at the left and correspond to those in Table 1 and Fig. 1. Each genotype table shows PB2 in the leftmost column, followed by PB1, PA, HA, NP, NA, M, and NS segments. Each row of a genotype table corresponds to a single plaque clone isolated from the indicated nasal-wash sample (n = 18 to 21). The red bars indicate segments derived from the wt parental strain, and the turquoise bars indicate segments derived from the var virus. White bars are shown where segments could not be typed unambiguously.
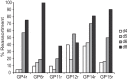
FIG 3
Prevalence of reassortant viruses in nasal washes collected from guinea pigs coinfected through two independent transmission events. The percentage of virus isolates that were found to carry reassortant genomes is plotted for each guinea pig and each time postinoculation that was examined in detail. Guinea pig ID numbers are shown as categories on the x axis and correspond to those in Fig. 1 and 2 and Table 1. The day postinoculation is indicated by the legend. The data shown correspond to genotypes displayed in Fig. 2.
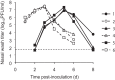
FIG 4
Nasal-wash titers of donor and recipient guinea pigs in a cotransmission model. Donor and recipient guinea pigs were cocaged in pairs at 24 h postinoculation of the donor animals. Guinea pig ID numbers are indicated on the left. Recipient no. 1 was paired with donor no. 2, no. 3 with no. 4, and no. 5 with no. 6. The viral titers in nasal washes of recipient animals are plotted with solid lines and symbols. The titers of donor animals are shown with dashed lines and open symbols. The limit of detection (50 PFU/ml) is indicated with a horizontal dashed line, and titers below the limit of detection were plotted as 45 PFU/ml.
FIG 5
Viral genotypes sampled from nasal washes of guinea pigs coinfected through transmission of multiple variant viruses from a single donor host. On the left of the vertical line, genotype tables are shown for three donor guinea pigs coinfected through intranasal administration of 104 PFU Pan/99wt and 104 PFU Pan/99var6 viruses. On the right of the line, genotype tables are shown for the corresponding recipient guinea pigs coinfected through contact exposure to the donor hosts with mixed infections. Recipient no. 1 was paired with donor no. 2, no. 3 with no. 4, and no. 5 with no. 6. The day postinoculation on which each nasal-wash sample was collected is indicated at the top. The guinea pig ID numbers are shown on the left or right for the donors and recipients, respectively, and correspond to those in Fig. 3. Each genotype table shows PB2 in the leftmost column, followed by the PB1, PA, HA, NP, NA, M, and NS segments. Each row of a genotype table corresponds to a single plaque clone isolated from the indicated nasal-wash sample (n = 17 to 21). The red bars indicate segments derived from the wt parental strain, and the turquoise bars indicate segments derived from the var virus. White bars are shown where segments could not be typed unambiguously. nd, time points at which virus was not detected in nasal washes of the indicated guinea pigs.
FIG 6
Prevalences of reassortant viruses in nasal washes collected from coinfected donor and recipient guinea pigs. The percentage of virus isolates that were found to carry reassortant genomes is plotted for each guinea pig and each time postinoculation examined. Guinea pig ID numbers are shown as categories on the x axis and correspond to those in Fig. 4 and 5. The day postinoculation is indicated by the legend. The data shown correspond to genotypes displayed in Fig. 5.
Similar articles
- Influenza A Virus Reassortment Is Limited by Anatomical Compartmentalization following Coinfection via Distinct Routes.
Richard M, Herfst S, Tao H, Jacobs NT, Lowen AC. Richard M, et al. J Virol. 2018 Feb 12;92(5):e02063-17. doi: 10.1128/JVI.02063-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29212934 Free PMC article. - Seasonal H3N2 and 2009 Pandemic H1N1 Influenza A Viruses Reassort Efficiently but Produce Attenuated Progeny.
Phipps KL, Marshall N, Tao H, Danzy S, Onuoha N, Steel J, Lowen AC. Phipps KL, et al. J Virol. 2017 Aug 10;91(17):e00830-17. doi: 10.1128/JVI.00830-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28637755 Free PMC article. - Intrahost dynamics of influenza virus reassortment.
Tao H, Steel J, Lowen AC. Tao H, et al. J Virol. 2014 Jul;88(13):7485-92. doi: 10.1128/JVI.00715-14. Epub 2014 Apr 16. J Virol. 2014. PMID: 24741099 Free PMC article. - Constraints, Drivers, and Implications of Influenza A Virus Reassortment.
Lowen AC. Lowen AC. Annu Rev Virol. 2017 Sep 29;4(1):105-121. doi: 10.1146/annurev-virology-101416-041726. Epub 2017 May 26. Annu Rev Virol. 2017. PMID: 28548881 Review. - Implications of segment mismatch for influenza A virus evolution.
White MC, Lowen AC. White MC, et al. J Gen Virol. 2018 Jan;99(1):3-16. doi: 10.1099/jgv.0.000989. Epub 2017 Dec 15. J Gen Virol. 2018. PMID: 29244017 Free PMC article. Review.
Cited by
- Influenza B Virus Infection Is Enhanced Upon Heterotypic Co-infection With Influenza A Virus.
Malausse N, van der Werf S, Naffakh N, Munier S. Malausse N, et al. Front Microbiol. 2021 Feb 25;12:631346. doi: 10.3389/fmicb.2021.631346. eCollection 2021. Front Microbiol. 2021. PMID: 33717023 Free PMC article. - Influenza A Virus Reassortment Is Limited by Anatomical Compartmentalization following Coinfection via Distinct Routes.
Richard M, Herfst S, Tao H, Jacobs NT, Lowen AC. Richard M, et al. J Virol. 2018 Feb 12;92(5):e02063-17. doi: 10.1128/JVI.02063-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29212934 Free PMC article. - Airborne Influenza A Is Detected in the Personal Breathing Zone of Swine Veterinarians.
O'Brien KM, Nonnenmann MW. O'Brien KM, et al. PLoS One. 2016 Feb 11;11(2):e0149083. doi: 10.1371/journal.pone.0149083. eCollection 2016. PLoS One. 2016. PMID: 26867129 Free PMC article. - Influenza Virus Reassortment Is Enhanced by Semi-infectious Particles but Can Be Suppressed by Defective Interfering Particles.
Fonville JM, Marshall N, Tao H, Steel J, Lowen AC. Fonville JM, et al. PLoS Pathog. 2015 Oct 6;11(10):e1005204. doi: 10.1371/journal.ppat.1005204. eCollection 2015 Oct. PLoS Pathog. 2015. PMID: 26440404 Free PMC article. - Evolution of pathogen tolerance and emerging infections: A missing experimental paradigm.
Seal S, Dharmarajan G, Khan I. Seal S, et al. Elife. 2021 Sep 21;10:e68874. doi: 10.7554/eLife.68874. Elife. 2021. PMID: 34544548 Free PMC article. Review.
References
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. doi:10.1126/science.1176225. - DOI - PMC - PubMed
- Li KS, Guan Y, Wang J, Smith GJ, Xu KM, Duan L, Rahardjo AP, Puthavathana P, Buranathai C, Nguyen TD, Estoepangestie AT, Chaisingh A, Auewarakul P, Long HT, Hanh NT, Webby RJ, Poon LL, Chen H, Shortridge KF, Yuen KY, Webster RG, Peiris JS. 2004. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430:209–213. doi:10.1038/nature02746. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources