Repbase Update, a database of repetitive elements in eukaryotic genomes - PubMed (original) (raw)
Repbase Update, a database of repetitive elements in eukaryotic genomes
Weidong Bao et al. Mob DNA. 2015.
Abstract
Repbase Update (RU) is a database of representative repeat sequences in eukaryotic genomes. Since its first development as a database of human repetitive sequences in 1992, RU has been serving as a well-curated reference database fundamental for almost all eukaryotic genome sequence analyses. Here, we introduce recent updates of RU, focusing on technical issues concerning the submission and updating of Repbase entries and will give short examples of using RU data. RU sincerely invites a broader submission of repeat sequences from the research community.
Keywords: Database; Repbase Reports; Repbase Update; RepbaseSubmitter; Transposable element.
Figures
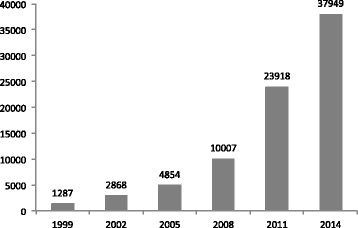
Fig. 1
Numbers of the entries in Repbase Update since 1999
Similar articles
- Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor.
Kohany O, Gentles AJ, Hankus L, Jurka J. Kohany O, et al. BMC Bioinformatics. 2006 Oct 25;7:474. doi: 10.1186/1471-2105-7-474. BMC Bioinformatics. 2006. PMID: 17064419 Free PMC article. - Repbase Update, a database of eukaryotic repetitive elements.
Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Jurka J, et al. Cytogenet Genome Res. 2005;110(1-4):462-7. doi: 10.1159/000084979. Cytogenet Genome Res. 2005. PMID: 16093699 Review. - VisualRepbase: an interface for the study of occurrences of transposable element families.
Tempel S, Jurka M, Jurka J. Tempel S, et al. BMC Bioinformatics. 2008 Aug 18;9:345. doi: 10.1186/1471-2105-9-345. BMC Bioinformatics. 2008. PMID: 18710569 Free PMC article. - Structural and sequence diversity of eukaryotic transposable elements.
Kojima KK. Kojima KK. Genes Genet Syst. 2020 Jan 30;94(6):233-252. doi: 10.1266/ggs.18-00024. Epub 2018 Nov 9. Genes Genet Syst. 2020. PMID: 30416149 Review. - Human transposable elements in Repbase: genomic footprints from fish to humans.
Kojima KK. Kojima KK. Mob DNA. 2018 Jan 4;9:2. doi: 10.1186/s13100-017-0107-y. eCollection 2018. Mob DNA. 2018. PMID: 29308093 Free PMC article. Review.
Cited by
- Integrated multi-approaches reveal unique metabolic mechanisms of Vestimentifera to adapt to deep sea.
Sun Q, Yuan Z, Sun Y, Sun L. Sun Q, et al. Microbiome. 2024 Nov 16;12(1):241. doi: 10.1186/s40168-024-01960-4. Microbiome. 2024. PMID: 39548600 Free PMC article. - The chromosome-level genome assembly and annotation of an invasive forest pest Obolodiplosis robiniae.
Huang L, Wang L, Sun HQ, Huai WX, Lin RZ, Wei SJ, Yao YX. Huang L, et al. Sci Data. 2024 Nov 14;11(1):1227. doi: 10.1038/s41597-024-04037-x. Sci Data. 2024. PMID: 39543130 Free PMC article. - Chromosome-level genome assembly of banaba (Lagerstroemia speciosa L.).
Wan Z, Zheng T, Cai M, Wang J, Pan H, Cheng T, Zhang Q. Wan Z, et al. Sci Data. 2024 Nov 14;11(1):1228. doi: 10.1038/s41597-024-04109-y. Sci Data. 2024. PMID: 39543103 Free PMC article. - Virulence perspective genomic research unlocks the secrets of Rhizoctonia solani associated with banded sheath blight in Barnyard Millet (Echinochloa frumentacea).
Patro TSSK, Palanna KB, Jeevan B, Tatineni P, Poonacha TT, Khan F, Ramesh GV, Nayak AM, Praveen B, Divya M, Anuradha N, Rani YS, Nagaraja TE, Madhusudhana R, Satyavathi CT, Prasanna SK. Patro TSSK, et al. Front Plant Sci. 2024 Oct 28;15:1457912. doi: 10.3389/fpls.2024.1457912. eCollection 2024. Front Plant Sci. 2024. PMID: 39529934 Free PMC article. - A Chromosome-Level Genome Assembly of Chiton Acanthochiton rubrolineatus (Chitonida, Polyplacophora, Mollusca).
Qu J, Lu X, Tu C, He F, Li S, Gu D, Wang S, Xing Z, Zheng L, Wang X, Wang L. Qu J, et al. Animals (Basel). 2024 Nov 4;14(21):3161. doi: 10.3390/ani14213161. Animals (Basel). 2024. PMID: 39518884 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources