Transcriptional Regulatory Circuits Controlling Brown Fat Development and Activation - PubMed (original) (raw)
Review
. 2015 Jul;64(7):2369-75.
doi: 10.2337/db15-0203. Epub 2015 Jun 7.
Affiliations
- PMID: 26050669
- PMCID: PMC4477361
- DOI: 10.2337/db15-0203
Review
Transcriptional Regulatory Circuits Controlling Brown Fat Development and Activation
Patrick Seale. Diabetes. 2015 Jul.
Abstract
Brown and beige adipose tissue is specialized for heat production and can be activated to reduce obesity and metabolic dysfunction in animals. Recent studies also have indicated that human brown fat activity levels correlate with leanness. This has revitalized interest in brown fat biology and has driven the discovery of many new regulators of brown fat development and function. This review summarizes recent advances in our understanding of the transcriptional mechanisms that control brown and beige fat cell development.
© 2015 by the American Diabetes Association. Readers may use this article as long as the work is properly cited, the use is educational and not for profit, and the work is not altered.
Figures
Figure 1
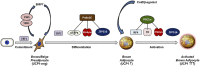
Transcriptional regulation of brown fat cell differentiation and activation. EBF2 marks committed brown preadipocytes and may regulate the commitment process from upstream stem cells. EWS/YBX1 regulates BMP7 production, which then acts in an autocrine manner to induce brown adipogenesis. EBF2, PRDM16, and ZFP516 specifically regulate the induction of brown fat–specific genes during the differentiation process. PRDM16 coactivates c/EBPβ, PPARγ, PPARα, thyroid receptor (TR), and ZFP516. Upon cold exposure/β-adrenergic agonist treatment, brown fat cells are activated to undergo thermogenesis and increase their expression of thermogenic genes. IRF4 plays a major role in this process through recruiting the PGC-1α coactivator. PGC-1α can also coactivate PPARs and TR to activate the transcription of thermogenic genes.
Figure 2
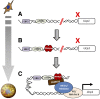
Model for transcriptional regulation of brown fat–selective genes through EBF2 and PRDM16. A: EBF2 binds to enhancer regions of brown fat genes at early time points during brown fat cell differentiation. B: Once bound, EBF2 facilitates the binding of PPARγ and other transcription factors to these enhancer regions. C: PRDM16 is recruited to these sites indirectly, likely through c/EBPβ, PPARγ, and/or ZFP516. PRDM16 binds and recruits the MED1/Mediator complex to facilitate enhancer/promoter looping and preinitiation complex (PIC) assembly. This action of PRDM16 is required to promote the efficient transcription of brown fat genes.
Similar articles
- MiR-27 orchestrates the transcriptional regulation of brown adipogenesis.
Sun L, Trajkovski M. Sun L, et al. Metabolism. 2014 Feb;63(2):272-82. doi: 10.1016/j.metabol.2013.10.004. Epub 2013 Oct 24. Metabolism. 2014. PMID: 24238035 - Brown and beige fat: development, function and therapeutic potential.
Harms M, Seale P. Harms M, et al. Nat Med. 2013 Oct;19(10):1252-63. doi: 10.1038/nm.3361. Epub 2013 Sep 29. Nat Med. 2013. PMID: 24100998 Review. - Peroxisome proliferator-activated receptors-α and -γ, and cAMP-mediated pathways, control retinol-binding protein-4 gene expression in brown adipose tissue.
Rosell M, Hondares E, Iwamoto S, Gonzalez FJ, Wabitsch M, Staels B, Olmos Y, Monsalve M, Giralt M, Iglesias R, Villarroya F. Rosell M, et al. Endocrinology. 2012 Mar;153(3):1162-73. doi: 10.1210/en.2011-1367. Epub 2012 Jan 17. Endocrinology. 2012. PMID: 22253419 - Transcriptional control of brown adipocyte development and thermogenesis.
Seale P. Seale P. Int J Obes (Lond). 2010 Oct;34 Suppl 1:S17-22. doi: 10.1038/ijo.2010.178. Int J Obes (Lond). 2010. PMID: 20935660 - Brown and Beige Fat: Molecular Parts of a Thermogenic Machine.
Cohen P, Spiegelman BM. Cohen P, et al. Diabetes. 2015 Jul;64(7):2346-51. doi: 10.2337/db15-0318. Epub 2015 Jun 7. Diabetes. 2015. PMID: 26050670 Free PMC article. Review.
Cited by
- TFAM expression in brown adipocytes confers obesity resistance by secreting extracellular vesicles that promote self-activation.
Fujii M, Setoyama D, Gotoh K, Dozono Y, Yagi M, Ikeda M, Ide T, Uchiumi T, Kang D. Fujii M, et al. iScience. 2022 Aug 10;25(9):104889. doi: 10.1016/j.isci.2022.104889. eCollection 2022 Sep 16. iScience. 2022. PMID: 36046191 Free PMC article. - β-arrestin-1 contributes to brown fat function and directly interacts with PPARα and PPARγ.
Wang C, Zeng X, Zhou Z, Zhao J, Pei G. Wang C, et al. Sci Rep. 2016 Jun 15;6:26999. doi: 10.1038/srep26999. Sci Rep. 2016. PMID: 27301785 Free PMC article. - Grape Seed Proanthocyanidins Improve White Adipose Tissue Expansion during Diet-Induced Obesity Development in Rats.
Pascual-Serrano A, Bladé C, Suárez M, Arola-Arnal A. Pascual-Serrano A, et al. Int J Mol Sci. 2018 Sep 5;19(9):2632. doi: 10.3390/ijms19092632. Int J Mol Sci. 2018. PMID: 30189642 Free PMC article. - Quercetin, Engelitin and Caffeic Acid of Smilax china L. Polyphenols, Stimulate 3T3-L1 Adipocytes to Brown-like Adipocytes Via β3-AR/AMPK Signaling Pathway.
Kong L, Zhang W, Liu S, Zhong Z, Zheng G. Kong L, et al. Plant Foods Hum Nutr. 2022 Dec;77(4):529-537. doi: 10.1007/s11130-022-00996-x. Epub 2022 Aug 20. Plant Foods Hum Nutr. 2022. PMID: 35986845 - Effects of maternal undernutrition during late pregnancy on the regulatory factors involved in growth and development in ovine fetal perirenal brown adipose tissue.
Yang H, Ma C, Zi Y, Zhang M, Liu Y, Wu K, Gao F. Yang H, et al. Anim Biosci. 2022 Jul;35(7):1010-1020. doi: 10.5713/ab.21.0199. Epub 2021 Sep 15. Anim Biosci. 2022. PMID: 34530507 Free PMC article.
References
- Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med 2013;19:1252–1263 - PubMed
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev 2004;84:277–359 - PubMed
- Nicholls DG, Rial E. A history of the first uncoupling protein, UCP1. J Bioenerg Biomembr 1999;31:399–406 - PubMed
- Enerbäck S, Jacobsson A, Simpson EM, et al. . Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 1997;387:90–94 - PubMed
- Lowell BB, S-Susulic V, Hamann A, et al. . Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature 1993;366:740–742 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources