Genomic comparison of closely related Giant Viruses supports an accordion-like model of evolution - PubMed (original) (raw)
Genomic comparison of closely related Giant Viruses supports an accordion-like model of evolution
Jonathan Filée. Front Microbiol. 2015.
Abstract
Genome gigantism occurs so far in Phycodnaviridae and Mimiviridae (order Megavirales). Origin and evolution of these Giant Viruses (GVs) remain open questions. Interestingly, availability of a collection of closely related GV genomes enabling genomic comparisons offer the opportunity to better understand the different evolutionary forces acting on these genomes. Whole genome alignment for five groups of viruses belonging to the Mimiviridae and Phycodnaviridae families show that there is no trend of genome expansion or general tendency of genome contraction. Instead, GV genomes accumulated genomic mutations over the time with gene gains compensating the different losses. In addition, each lineage displays specific patterns of genome evolution. Mimiviridae (megaviruses and mimiviruses) and Chlorella Phycodnaviruses evolved mainly by duplications and losses of genes belonging to large paralogous families (including movements of diverse mobiles genetic elements), whereas Micromonas and Ostreococcus Phycodnaviruses derive most of their genetic novelties thought lateral gene transfers. Taken together, these data support an accordion-like model of evolution in which GV genomes have undergone successive steps of gene gain and gene loss, accrediting the hypothesis that genome gigantism appears early, before the diversification of the different GV lineages.
Keywords: duplication; evolution; genome; giant virus; lateral gene transfer; mobile genetic elements.
Figures
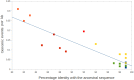
Figure 1
Number of genomic events per kb for each GVs with respect to the percentage of nucleotide similarities of the four taxonomic genes with the ancestral sequences. GVs are represented by squares: orange for Micromonas viruses, red for Ostreococcus viruses, green for Chlorella viruses, yellow for megaviruses, and blue for mimiviruses. The blue line represents the regression (_r_2 = 0.81).
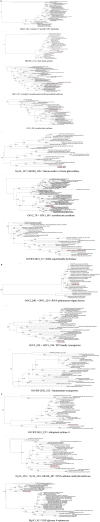
Figure 2
Genomic map of the megaviruses and the mimiviruses describing the different genomic mutations that have occurred after the divergence with their last common ancestor genome. Symbols below the line indicated gene losses (dots for gene losses, arrows for excision of mobile genetic elements and stars for translocation) and symbols above the line indicate gene gains (squares for duplications, triangle for LGTs and arrows for insertion of mobile genetic elements). Interrogation marks represent ambiguous cases or presence of orphans. Squares and dots in red indicate genomic events of genes belonging to paralogous families. The trees represent the whole genome phylogeny, the numbers above and below the terminal branches indicate the amount of gene gain and loss for each virus. The scale bar represents the equivalent of 100 kb.
Figure 3
Genomic map of the Chlorella and Ostreococcus Phycodnaviruses describing the different genomic mutations that have occurred after the divergence with their last common ancestor. Symbols below the line indicated gene losses (dots for gene losses, arrows for excision of mobile genetic elements) and symbols above the line indicate gene gains (squares for duplications, triangle for LGTs and arrows for insertion of mobile genetic elements). Interrogation marks represent ambiguous cases or presence of orphans. Squares and dots in red indicate genomic events of genes belonging to paralogous families. The trees represent the whole genome phylogeny, the numbers above and below the terminal branches indicate the amount of gene gain and loss for each virus. The scale bar represents the equivalent of 100 kb.
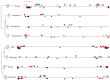
Figure 4
Phylogenies of different cases of LGT encountered in GVs. (A) LGT from diverse Bacteria and Eukarya (B) LGT from the hosts. (C) LGT from ambiguous sources. Viral sequences have been framed in red. The numbers beside each node indicate the value of the SH-like statistical test.
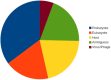
Figure 5
Taxonomic sources of the LGTs.
Similar articles
- Mimiviridae: clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family.
Yutin N, Colson P, Raoult D, Koonin EV. Yutin N, et al. Virol J. 2013 Apr 4;10:106. doi: 10.1186/1743-422X-10-106. Virol J. 2013. PMID: 23557328 Free PMC article. - Evolution of the Large Nucleocytoplasmic DNA Viruses of Eukaryotes and Convergent Origins of Viral Gigantism.
Koonin EV, Yutin N. Koonin EV, et al. Adv Virus Res. 2019;103:167-202. doi: 10.1016/bs.aivir.2018.09.002. Epub 2018 Nov 10. Adv Virus Res. 2019. PMID: 30635076 Review. - Evolutionary genomics of nucleo-cytoplasmic large DNA viruses.
Iyer LM, Balaji S, Koonin EV, Aravind L. Iyer LM, et al. Virus Res. 2006 Apr;117(1):156-84. doi: 10.1016/j.virusres.2006.01.009. Epub 2006 Feb 21. Virus Res. 2006. PMID: 16494962 Review. - Evolution and Phylogeny of Large DNA Viruses, Mimiviridae and Phycodnaviridae Including Newly Characterized _Heterosigma akashiwo Viru_s.
Maruyama F, Ueki S. Maruyama F, et al. Front Microbiol. 2016 Nov 30;7:1942. doi: 10.3389/fmicb.2016.01942. eCollection 2016. Front Microbiol. 2016. PMID: 27965659 Free PMC article. - Convergent mechanisms of genome evolution of large and giant DNA viruses.
Filée J, Chandler M. Filée J, et al. Res Microbiol. 2008 Jun;159(5):325-31. doi: 10.1016/j.resmic.2008.04.012. Epub 2008 May 10. Res Microbiol. 2008. PMID: 18572389
Cited by
- Adaptation by copy number variation in monopartite viruses.
Bayer A, Brennan G, Geballe AP. Bayer A, et al. Curr Opin Virol. 2018 Dec;33:7-12. doi: 10.1016/j.coviro.2018.07.001. Epub 2018 Jul 14. Curr Opin Virol. 2018. PMID: 30015083 Free PMC article. Review. - Functional Genomic Analyses Reveal an Open Pan-genome for the Chloroviruses and a Potential for Genetic Innovation in New Isolates.
Rodrigues RAL, Queiroz VF, Ghosh J, Dunigan DD, Van Etten JL. Rodrigues RAL, et al. J Virol. 2022 Jan 26;96(2):e0136721. doi: 10.1128/JVI.01367-21. Epub 2021 Oct 20. J Virol. 2022. PMID: 34669449 Free PMC article. - Widespread endogenization of giant viruses shapes genomes of green algae.
Moniruzzaman M, Weinheimer AR, Martinez-Gutierrez CA, Aylward FO. Moniruzzaman M, et al. Nature. 2020 Dec;588(7836):141-145. doi: 10.1038/s41586-020-2924-2. Epub 2020 Nov 18. Nature. 2020. PMID: 33208937 - Evolution of giant pandoravirus revealed by CRISPR/Cas9.
Bisio H, Legendre M, Giry C, Philippe N, Alempic JM, Jeudy S, Abergel C. Bisio H, et al. Nat Commun. 2023 Jan 26;14(1):428. doi: 10.1038/s41467-023-36145-4. Nat Commun. 2023. PMID: 36702819 Free PMC article. - Pangenomic Analysis of Nucleo-Cytoplasmic Large DNA Viruses. I: The Phylogenetic Distribution of Conserved Oxygen-Dependent Enzymes Reveals a Capture-Gene Process.
Campillo-Balderas JA, Lazcano A, Cottom-Salas W, Jácome R, Becerra A. Campillo-Balderas JA, et al. J Mol Evol. 2023 Oct;91(5):647-668. doi: 10.1007/s00239-023-10126-z. Epub 2023 Aug 1. J Mol Evol. 2023. PMID: 37526693 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources