CCHamide-2 Is an Orexigenic Brain-Gut Peptide in Drosophila - PubMed (original) (raw)
. 2015 Jul 13;10(7):e0133017.
doi: 10.1371/journal.pone.0133017. eCollection 2015.
Frank Hauser 1, Kim F Rewitz 2, Shu Kondo 3, Alexander F Engelbrecht 1, Anders K Didriksen 1, Suzanne R Schjøtt 1, Frederikke E Sembach 1, Shizhong Li 1, Karen C Søgaard 1, Leif Søndergaard 1, Cornelis J P Grimmelikhuijzen 1
Affiliations
- PMID: 26168160
- PMCID: PMC4500396
- DOI: 10.1371/journal.pone.0133017
CCHamide-2 Is an Orexigenic Brain-Gut Peptide in Drosophila
Guilin R Ren et al. PLoS One. 2015.
Abstract
The neuroendocrine peptides CCHamide-1 and -2, encoded by the genes ccha1 and -2, are produced by endocrine cells in the midgut and by neurons in the brain of Drosophila melanogaster. Here, we used the CRISPR/Cas9 technique to disrupt the ccha1 and -2 genes and identify mutant phenotypes with a focus on ccha-2 mutants. We found that both larval and adult ccha2 mutants showed a significantly reduced food intake as measured in adult flies by the Capillary Feeding (CAFE) assay (up to 72% reduced food intake compared to wild-type). Locomotion tests in adult flies showed that ccha2 mutants had a significantly reduced locomotor activity especially around 8 a.m. and 8 p.m., where adult Drosophila normally feeds (up to 70% reduced locomotor activity compared to wild-type). Reduced larval feeding is normally coupled to a delayed larval development, a process that is mediated by insulin. Accordingly, we found that the ccha2 mutants had a remarkably delayed development, showing pupariation 70 hours after the pupariation time point of the wild-type. In contrast, the ccha-1 mutants were not developmentally delayed. We also found that the ccha2 mutants had up to 80% reduced mRNA concentrations coding for the Drosophila insulin-like-peptides-2 and -3, while these concentrations were unchanged for the ccha1 mutants. From these experiments we conclude that CCHamide-2 is an orexigenic peptide and an important factor for controlling developmental timing in Drosophila.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Fig 1. Nucleotide sequences and corresponding amino acid sequences around the deletions in two ccha1 (A) and two ccha2 (B) mutants.
In the wild-type these nucleotide sequences code for the unprocessed CCHamide peptides, which are shown in red at the top of each panel. The black arrows in these red lines at the top indicate the initial cleavage steps in each prohormone, catalyzed by prohormone convertase [38]. A. Parts of the DNA sequences from the two ccha1 mutants (ccha1 SK4 and ccha1 SK8) and the corresponding wild-type DNA sequence coding for CCHamide-1. Mutant ccha1 SK4 lacks 5 base pairs (bp), while mutant ccha SK8 lacks 13 bp. Both deletions lead to a frameshift, so that no intact CCHamide-1 peptide can be produced. For example, while in the wild-type the two cysteine residues (underlined) form a cystine bridge, such ring structure can not be formed in the mutant peptides, because a second cysteine residue is lacking. Furthermore, while in the wild-type processing occurs between the KR and S amino acid sequence (arrow), followed by a conversion of the C-terminal G residue into a C-terminal amide [38], such posttranslational processings can not occur in the two mutants, due to the lack of the GKR amino acid sequence at these positions. The mutations, therefore, result in nonfunctional peptides that only have the N-terminal amino acid residues in common with wild-type CCHamide-1. B. Parts of the DNA sequences from ccha2 mutants and their corresponding wild-type DNA sequences coding for CCHamide-2. The two mutants have identical 10 bp deletions that, again, cause a frameshift in the reading frame, resulting in the loss of the cystine bridge and the appropriate processing sites to yield functional peptides. Furthermore, the two mutants have a premature stop codon (TGA).
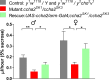
Fig 2. The capillary feeding (CAFE) assay for ccha2 mutant male and female adult flies.
Each data point was an average from the results obtained from 10 tubes containing 4 flies each. The experiments were repeated five times, each time with fresh animals. The controls are indicated by black bars, the mutants (ccha2 SK1/ccha2 SK3) by red bars, while the rescued mutants are indicated by green bars. The mutant male flies have 30% feeding activity left compared to the controls (n = 5; t-test, ** p≤0.01), while the mutant female flies have 37% feeding activity left compared to the controls (n = 5; t-test, * p≤0.5). The rescued male and female ccha2 mutants more than doubled their feeding activities compared to the ccha2 null mutants (*p≤0.5). The vertical bars represent S.E.M.
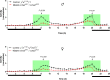
Fig 3. Circadian activities of the control (black lines) and CCHamide-2 mutants (red lines).
The activities were measured using a Drosophila activity monitor that monitors one-dimensional locomotion of single flies. Light is switched on at 8 a.m. and switched off at 8 p.m. The upper panel gives the activities of 6-d old male, and the lower panel of 6-d old female flies. The data points represent the average of three independent experiments containing 32 flies each (n = 3). The vertical bars represent S.E.M. When no vertical bars are visible, they are smaller than the symbols used. The green areas highlight time periods around 8 a.m. and 8 p.m., where the activity differences between mutants and wild-type were especially significant. These periods coincide with the normal feeding periods of wild-type Drosophila [25]. The arrows indicate significant activity differences between mutants and controls at 8 a.m. and 8 p.m. P-values are between p≤0.001 and p≤0.05.
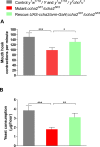
Fig 4. Larval feeding assays for control flies (black bars), ccha2 null mutants (red bars) are rescued ccha2 mutants (green bars).
A. Larval feeding assay measuring the frequency of mouth hook contractions of third instar larvae feeding on agar covered with a 2% yeast solution. The ccha2 null mutants have 63% of their feeding activity left compared to the controls (n = 5; t-test. *** p≤0.001). The rescued mutants restored their feeding activity to a level which is 86% of the control activity. The difference between ccha2 null mutants and rescued mutants is significant (t-test, * p≤0.5). B. A different larval feeding assay, measuring the amount of ingested color-labelled yeast per hour. The ccha2 null mutants have 43% of their feeding activities left compared to controls (n = 5; t-test, *** p≤0.001). The rescued mutants restored their feeding activity to 78% of the controls. The difference between ccha2 null and rescued mutants is significant (t-test, ** p≤0.01).
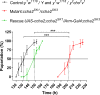
Fig 5. Pupariation time points of ccha2 mutants compared to control.
The horizontal line parallel to the abscissa indicates 50% of the animals having undergone pupariation. The vertical stippled lines indicate the time points, where 50% of the experimental animals have pupariated. Control animals (indicated by a black line) pupariated (pupal stage P-2) at 132 hrs after egg laying Homozygous mutants (indicated by a red line) pupariated at 202 hrs after egg laying and were, therefore, 70 hrs delayed compared to controls. Furthermore, ccha2 mutants rescued by re-introducing the ccha2 gene (indicated by a green line) pupariated at 148 hrs and were, thus rescued by 80%. The data points represent the average of five independent experiments, containing 15–25 animals each. The vertical bars represent S.E.M. The differences between control and ccha2 mutants, and between ccha2 mutants and rescued mutants are stastically significant (one-way ANOVA test, p≤0.001).
Fig 6. qPCR of Drosophila insulin-like peptide (DILP) gene expressions in third instar larvae and pupae of ccha1 and -2 null mutants and wild-type animals.
Control animals are indicated by black bars, ccha1 mutants are indicated by white bars, ccha2 mutants are indicated by red bars. The vertical bars represent S.E.M. (n = 3). Thirty animals were used in each measurement (2 technical replicates; 3 biological replicates). A. In larval ccha2 mutants, dilp2 gene expression is reduced by about 50% (t-test, *** p≤0.001), while in larval ccha1 mutants there is no reduction compared to wild-type. B. In pupal ccha2 mutants (pupal stage P-5), dilp2 gene expression is reduced to 35% of the wild-type values (t-test, *** p≤0.001), while there is no reduction in ccha1 mutants. C. In larval ccha2 mutants, dilp3 gene expression is reduced to 20% of the wild-type values (t-test, *** p≤0.001), while there is no such downregulation in ccha1 mutants. D. In pupal ccha2 mutants (stage P-5), the dilp3 gene expression is downregulated to about 50% of the wildtype values (t-test, *** p≤0.001), while there is no significant downregulation in the pupal ccha1 mutants.
Fig 7. Wing size and adult fly weight in ccha2 disruption mutants compared to wild-type.
Wild-type animals are indicated by black bars, ccha2 mutants are indicated by red bars. Vertical bars represent S.E.M. (n = 30). A. The wing surface of male ccha2 mutants is 22.7% reduced compared to wild-types (n = 30; student t-test, *** p≤0.001). B. The wing surface of female mutants is 15.2% reduced compared to wild-types (n = 30; student t-test *** p≤0.0001). C. There is no significant weight difference between male ccha2 mutants and wild-types (n = 100). D. There is no significant weight difference between female ccha2 mutants and wild-types (n = 100).
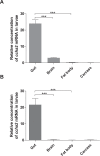
Fig 8. Expresion of the ccha2 gene in different organs of mid third instar D. melanogaster larvae (92 hrs after egg laying).
Two primer sets were used for qPCR: One primer set previously applied by us [13] and one primer set used by Sano et al. [18] (S1 Table). (A) qPCR results using the primer set described by us [13]. (B) qPCR results using the primer set described by Sano et al. [18]. It is clear from both experiments that the gut is the major source of ccha2 mRNA, while the fat body is virtually devoid of ccha2 mRNA (n = 3; student t-test *** p≤0.001).
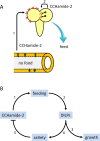
Fig 9. Hypothetical model for the actions of CCHamide-2 in D. melanogaster.
A. When the lumen of the midgut (lower part of Fig 8A) is devoid of nutrients, the CCHamide-2 containing endocrine cells of the gut wall (highlighted in green) signal this information to the brain by releasing CCHamide-2 into the circulation (arrow 1). After binding to its brain receptors, CCHamide-2 induces foraging and feeding behavior. In addition to this long-distance CCHamide-2 signaling pathway, there is a short distance CCHamide-2 signaling pathway (arrow 2), where a small group of CCHamide-2 neurons in the brain (highlighted in green) also innervate the motor circuits underlying foraging and feeding. We hypothesize that these neurons might perhaps directly monitor the nutrients in the circulation. B. A flow diagram of the proposed sequence of events after CCHamide-2 has induced feeding (step 1; see also Fig 8A). Feeding induces the release of DILPs (Step 2; see refs 26, 31, 32). DILPs stimulate growth (step 3; see refs 26, 28), but also induce satiety (step 4; see ref 30, 31, 33). It is assumed that satiety blocks the release of CCHamide-2 and other orexigenic neuropeptides.
Similar articles
- Expression patterns of the Drosophila neuropeptide CCHamide-2 and its receptor may suggest hormonal signaling from the gut to the brain.
Li S, Torre-Muruzabal T, Søgaard KC, Ren GR, Hauser F, Engelsen SM, Pødenphanth MD, Desjardins A, Grimmelikhuijzen CJ. Li S, et al. PLoS One. 2013 Oct 2;8(10):e76131. doi: 10.1371/journal.pone.0076131. eCollection 2013. PLoS One. 2013. PMID: 24098432 Free PMC article. - The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of Drosophila melanogaster.
Sano H, Nakamura A, Texada MJ, Truman JW, Ishimoto H, Kamikouchi A, Nibu Y, Kume K, Ida T, Kojima M. Sano H, et al. PLoS Genet. 2015 May 28;11(5):e1005209. doi: 10.1371/journal.pgen.1005209. eCollection 2015 May. PLoS Genet. 2015. PMID: 26020940 Free PMC article. - More Drosophila enteroendocrine peptides: Orcokinin B and the CCHamides 1 and 2.
Veenstra JA, Ida T. Veenstra JA, et al. Cell Tissue Res. 2014 Sep;357(3):607-21. doi: 10.1007/s00441-014-1880-2. Epub 2014 May 22. Cell Tissue Res. 2014. PMID: 24850274 - Drosophila neuropeptides in regulation of physiology and behavior.
Nässel DR, Winther AM. Nässel DR, et al. Prog Neurobiol. 2010 Sep;92(1):42-104. doi: 10.1016/j.pneurobio.2010.04.010. Epub 2010 May 4. Prog Neurobiol. 2010. PMID: 20447440 Review. - Leucokinin and Associated Neuropeptides Regulate Multiple Aspects of Physiology and Behavior in Drosophila.
Nässel DR. Nässel DR. Int J Mol Sci. 2021 Feb 16;22(4):1940. doi: 10.3390/ijms22041940. Int J Mol Sci. 2021. PMID: 33669286 Free PMC article. Review.
Cited by
- Neural circuit mechanisms encoding motivational states in Drosophila.
Lee SS, Wu MN. Lee SS, et al. Curr Opin Neurobiol. 2020 Oct;64:135-142. doi: 10.1016/j.conb.2020.05.002. Epub 2020 Jun 18. Curr Opin Neurobiol. 2020. PMID: 32563845 Free PMC article. Review. - Effect of Dietary Restriction on Gut Microbiota and Brain-Gut Short Neuropeptide F in Mud Crab, Scylla paramamosain.
Bao C, Yang Y, Ye H. Bao C, et al. Animals (Basel). 2024 Aug 20;14(16):2415. doi: 10.3390/ani14162415. Animals (Basel). 2024. PMID: 39199949 Free PMC article. - The Role of Peptide Hormones in Insect Lipid Metabolism.
Toprak U. Toprak U. Front Physiol. 2020 May 7;11:434. doi: 10.3389/fphys.2020.00434. eCollection 2020. Front Physiol. 2020. PMID: 32457651 Free PMC article. Review. - Pleiotropic fitness effects of a Drosophila odorant-binding protein.
Mokashi SS, Shankar V, Johnstun JA, Mackay TFC, Anholt RRH. Mokashi SS, et al. G3 (Bethesda). 2023 Feb 9;13(2):jkac307. doi: 10.1093/g3journal/jkac307. G3 (Bethesda). 2023. PMID: 36454098 Free PMC article. - Interorgan communication through peripherally derived peptide hormones in Drosophila.
Okamoto N, Watanabe A. Okamoto N, et al. Fly (Austin). 2022 Dec;16(1):152-176. doi: 10.1080/19336934.2022.2061834. Fly (Austin). 2022. PMID: 35499154 Free PMC article. Review.
References
- Zdárek J, Nachman RJ, Denlinger DL (2000) Parturition hormone in the tsetse Glossina morsitans: activity in reproductive tissues from other species and response of tsetse to identified neuropeptides and other neuroactive compounds. J Insect Physiol 46: 213–219. - PubMed
- Roller L, Yamanaka N, Watanabe K, Daubnerová I, Zitnan D, Kataoka H et al. (2008) The unique evolution of neuropeptide genes in the silkworm Bombyx mori . Insect Biochem Mol Biol 38: 1147–1157. - PubMed
- Hansen KK, Hauser F, Williamson M, Weber SB, Grimmelikhuijzen CJP (2011) The Drosophila genes CG14593 and CG30106 code for G-protein-coupled receptors specifically activated by the neuropeptides CCHamide-1 and CCHamide-2. Biochem Biophys Res Commun: 404: 184–189. 10.1016/j.bbrc.2010.11.089 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases