Determination of cell uptake pathways for tumor inhibitor lysyl oxidase propeptide - PubMed (original) (raw)
Determination of cell uptake pathways for tumor inhibitor lysyl oxidase propeptide
Gokhan Baris Ozdener et al. Mol Oncol. 2016 Jan.
Abstract
The lysyl oxidase propeptide (LOX-PP) is derived from pro-lysyl oxidase (Pro-LOX) by extracellular biosynthetic proteolysis. LOX-PP inhibits breast and prostate cancer xenograft tumor growth and has tumor suppressor activity. Although, several intracellular targets and molecular mechanisms of action of LOX-PP have been identified, LOX-PP uptake pathways have not been reported. Here we demonstrate that the major uptake pathway for recombinant LOX-PP (rLOX-PP) is PI3K-dependent macropinocytosis in PWR-1E, PC3, SCC9, MDA-MB-231 cell lines. A secondary pathway appears to be dynamin- and caveola dependent. The ionic properties of highly basic rLOX-PP provide buffering capacity at both high and low pHs. We suggest that the buffering capacity of rLOX-PP, which serves to limit endosomal acidification, sustains PI3K-dependent macropinocytosis in endosomes which in turn is likely to facilitate LOX-PP endosomal escape into the cytoplasm and its observed interactions with cytoplasmic targets and nuclear uptake.
Keywords: Endocytosis; Lysyl oxidase; Macropinocytosis; Propeptide; Tumor suppressor.
Copyright © 2015 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures
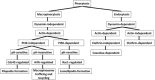
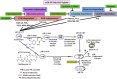
Figure 1
Scheme for the pinocytosis pathways evaluated.
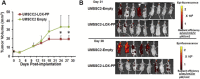
Figure 2
rLOX‐PP inhibits orthotopic growth and metastasis of human UMSCC2 oral cancer cells. UMSCC2 oral cancer cells expressing DsRed (Bais et al., 2015a) were transduced with a lentivirus expression vector for rLOX‐PP (n = 8) or empty vector (n = 9). Cells were respectively orthotopically injected into nude mice at 500,000 cells per mouse. (A) Caliper measurements were carried out at intervals and tumor volumes calculated. (B) In vivo imaging of tumor cells in mice was conducted at intervals. Data indicate that rLOX‐PP expression inhibited both primary tumor growth and formation of metastases. *p < 0.05, two way ANOVA with Bonferroni post hoc analysis.
Figure 3
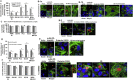
Time‐dependent cell uptake of rLOX‐PP‐Atto565 by a variety of cell lines. (A) Cell lines were incubated with 0.2 μM rLOX‐PP‐Atto565 and uptake was determined as a function of time by flow cytometry. Data are means of samples analyzed in triplicate ± SD, and experiments were performed at least twice. (B) Live cell imaging of selected cell lines by confocal microscopy after three hours further supports that rLOX‐PP‐Atto‐565 (red) was internalized. Both cytoplasmic and apparent nuclear localization was observed. Nuclei were stained with Hoechst 33342.
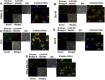
Figure 4
Co‐localization of rLOX‐PP‐Atto565 and 10 kDa dextran‐Bodipy‐fl. Cells were incubated with 10 μM 10 kDa dextran‐Bodipy‐fl (green) and 0.2 μM rLOX‐PP‐Atto565 (red) for 3 h before imaging by confocal microscopy. Confocal microscope images were formatted in split Z‐stacks on the left and merged on the right for each cell line: (A) PWR‐1E, (B) DU145, (C) PC3, (D) SCC9 and (E) MDA‐MC‐231. Corresponding dichromatic (DIC) images of cells are shown for each cell line.
Figure 5
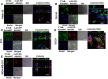
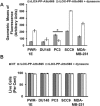
Inhibition of rLOX‐PP‐Atto565 uptake in the presence or absence of cytochalasin D (A–C) or LY294002 (D–F). (A) After 3 h rLOX‐PP‐Atto565 uptake was quantified by flow cytometry in absence (grey) and presence of 1.5 μM cytochalasin D (white); n = 3; *, two‐tailed p‐value > 0.0002. SCC9 cells were pre‐incubated on ice for 30 min in the absence (B‐1a and B‐1b) or presence (B‐2) of cytochalasin D. rLOX‐PP‐Atto565 was added in the presence or absence of cytochalasin D for an additional 15 min on ice, and then incubated at 37 °C for 15 min in the 5% CO2 incubator. Cells were stained for F‐actin (green) and DNA (blue) and indicate that 1.5 μM cytochalasin D disrupted actin filaments, as expected. Merged Z‐series images of without cytochalasin (B‐1a and B1b) and with cytochalasin D (B‐2) treatment were reconstructed with the LSM image viewer software. (C) The LIVE/DEAD® Fixable Near‐IR stain assay determined the percentage of live cells in each sample. (NT, dark gray bars; non treated control cells; rLOX‐PP‐Atto565, light gray bars; rLOX‐PP‐Atto565 + cytochalasin D, white bars). (D) rLOX‐PP‐Atto565 uptake was quantified by flow cytometry in the absence (gray bar) and in the presence of 100 μM LY294002 (white bar). Data are means ± SD; n = 3; *, two‐tailed p‐value>0.0006. (E) SCC9 cells were treated with rLOX‐PP‐Atto565 (red) for 15 min and stained for F‐actin (green) and DNA (blue) in absence and presence of 100 μM LY294002. Merged Z‐series images of B without LY294002 (above) and with LY294002 (below) treatment were reconstructed with image J software. 3 dimensional images from various angles show cup formation that open to plasma membrane (F). The LIVE/DEAD® Fixable Near‐IR stain assay was employed to determine the percentage of live cells in each cell lines; NT, dark gray bars, non‐treated control cells; rLOX‐PP‐Atto565, light gray bars; LY294002 plus rLOX‐PP‐Atto565, white bars.
Figure 6
Co‐localization of rLOX‐PP‐Atto565 and CTxB‐Alexa647 in various cell lines. Cells were incubated with CtxB‐Alexa647 (652/668 nm‐magenta) and rLOX‐PP‐Atto565 (563/592 nm‐red) for 3 h before imaging. Confocal microscope pictures were formatted in split Z‐stacks on the left and merged on the right for each cell line. PWR‐1E (A), DU145 (B), PC3 (C), SCC9 (D) and MDA‐MD‐231(E) cell lines were listed; (DIC), dichromatic image of cells. Various levels of co‐localization of rLOX‐PP‐Atto565 with CTxB‐Alexa647 in PWR‐1E, PC3, and MDA‐MC‐231 cell lines confirm that rLOX‐PP‐Atto565 enters the cell through a caveola‐mediated pathway.
Figure 7
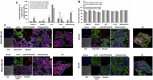
Inhibition of caveolae‐mediated rLOX‐PP‐Atto565 and CtxB‐Alexa647 uptake by siRNA knockdown of the primary caveolae protein caveolin‐1. (A) After transfection, cells were serum starved for 12 h followed by incubation with or without rLOX‐PP‐Atto565 or CtxB‐Alexa647 for an additional 3 h rLOX‐PP‐Atto565 (solid gray and white) and CtxB‐Alexa 647 (dashed gray and white) uptake were quantified by flow cytometry with (white) and without of CAV‐1 knockdown (gray); Data are means ± SD, n = 3.; *, two‐tailed p‐value < 0.005 (B) The LIVE/DEAD® Fixable Near‐IR stain assay was employed to determine the percentage of live cells in each sample. (NT, dark solid or dashed gray bars; non‐treated control cells; rLOX‐PP‐Atto565 or CtxB‐Alexa647, solid or dashed light gray bars; rLOX‐PP‐Atto565 or CtxB‐Alexa647 + caveolin‐1 siRNA knockdown, solid or dashed light white bars). Data are means ± SD, n = 3. (C) MDA‐MB‐231 (left) and SCC9 cells (right) were transfected with either caveolin‐1 siRNA or control siRNA. 60 h after transfection, cells were serum starved for 12 h followed by incubation with or without rLOX‐PP‐Atto565 (red) for an additional 15 min on ice, and then incubated at 37 °C for 15 min in the 5% CO2 incubator. Cells then were fixed, permeabilized and stained for F‐actin (green), DNA (blue) and total caveolin‐1 (magenta) or phospho caveolin‐1 (magenta). Merged Z‐series images with rLOX‐PP‐Atto565 (above) and without rLOX‐PP‐Atto565 (below) treatment were reconstructed with Zen Black Edition software. 3D images were constructed with Image J software.
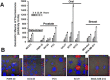
Figure 8
siRNA knockdown of the caveolin‐1 protein expression and the correlation of phospho caveolin‐1 protein expression level in cells incubated in serum, serum starved or serum starved and rLOX‐PP treated cells. (A) For confirmation of siRNA knockdown of the caveolin‐1 protein, cells were transfected under similar conditions with either control (non‐silencing) siRNA or siRNA directed against caveolin‐1. Maximum reduction of caveolin‐1 due to specific siRNA transfection was observed 75 h post‐transfection. Sixty hours after transfection, cells were serum starved for 12 h followed by incubation with or without rLOX‐PP for an additional 3 h. Cells extracts were subjected to SDS PAGE. Cell extract samples were probed with phosphor‐caveolin‐1, total caveolin‐1 and band intensity against beta actin was quantified and relative phospho‐CAV‐1 protein expression was determined. This experiment was performed at least three times with the same outcomes. Data are means ± SD; n = 3; *, two‐tailed p‐value <0.0001. (B) MDA‐MB‐231 (left) and SCC9 cells (right) were transfected with either CAV‐1 siRNA or control siRNA. After transfection, cells were serum starved for 12 h followed by incubation with or without rLOX‐PP‐Atto565 (red) for an additional 15 min on ice, and then incubated at 37 °C for 15 min in the 5% CO2 incubator. Cells then were fixed, permeabilized and stained for F‐actin (green), DNA (blue) and total caveolin‐1 (magenta) or –phospho caveolin‐1 (magenta). Merged images with rLOX‐PP‐Atto565 (above) and without rLOX‐PP‐Atto565 (below) treatment were reconstructed with Zen Black Edition software.
Figure 9
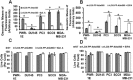
Co‐localization of rLOX‐PP‐Atto565 and transferrin‐FITC in various cell lines and inhibition of rLOX‐PP‐Atto565 (A–E), and 10 kDa dextran‐Bodipy‐fl and transferrin‐FITC uptake in presence or absence of dynasore in PC3 cells at 3 h (F). Cells were incubated with FITC‐transferrin (green) and rLOX‐PP‐Atto565 (red) for 15 min on ice, and then 30 min at 37 °C before imaging. Nuclei were stained with Hoechst 33342 (blue). Confocal microscope images were formatted in split Z‐stacks on the left and merged on the right for each cell lines. PWR‐1E (A), DU145 (B), PC3 (C), SCC9 (D) and MDA‐MC‐231(E) cell lines were listed. Dichromatic (DIC) images are also shown. Transferrin (green) and rLOX‐PP‐Atto565 (red) co‐localization was observed only in PC3 cells and confirms that rLOX‐PP‐Atto565 enters the PC3 cells through a clathrin‐mediated pathway. In (F), uptake of transferrin‐FITC was assessed after 30 min, while that of rLOX‐PP‐Atto565 and 10 kDa dextran‐Bodipy‐fl were assessed after three hours, in the presence or absence of dynasore in PC3 cells with or without quenching of extracellular fluorescence using a Zeiss Axiovert 100M inverted fluorescence microscope.
Figure 10
Inhibition of clathrin‐mediated uptake of rLOX‐PP‐Atto565 in presence or absence of dynasore. (A) After 3 h, rLOX‐PP‐Atto565 uptake quantified with flow cytometry in absence (gray bar) and presence of 100 μM dynasore (white bar); *, significant inhibition of uptake in PC3 cells only (p < 0.000125; n = 3). Up‐regulation occurred in all other cell lines. (B) The LIVE/DEAD® Fixable Near‐IR stain assay was employed to determine the percentage of live cells. NT (dark gray bars), non‐treated control cells; rLOX‐PP‐Atto565 (light gray bars); rLOX‐PP‐Atto565 plus dynasore (white bars). Data are means ± SD, n = 3.
Figure 11
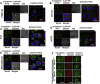
rLOX‐PP‐Atto565 uptake in bafilomycin A1 and EIPA treated cells. Cells were pre‐treated in the presence or absence of bafilomycin A1 (A) or EIPA (B), and rLOX‐PP‐Atto‐565 uptake was measured by flow cytometry after 30 min of incubation. Bafilomycin decreased rLOX‐PP‐Atto565 uptake after 30 min. Data are means ± SD, n = 3; *, p < 0.005. rLOX‐PP uptake was inhibited by bafilomycin A, while it was stimulated by EIPA in all cell lines except possibly DU145 cells. In C and D, the LIVE/DEAD® Fixable Near‐IR stain flow cytometry assay was used to determine the percentage of live cells. NT (dark gray bars), non‐treated control cells; rLOX‐PP‐Atto565 (light gray bars), bafilomycin A‐ or EIPA‐ + rLOX‐PP‐Atto565 (white bars).
Figure 12
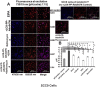
Internalized rLOXPP‐Atto647N increased endosomal pH from 6.77 to 6.85. In (A), SCC9 cells were maintained in the recording buffer at pH 7.15 for steady state extracellular pH and incubated with Lysosensor Yellow/Blue dextran ± rLOX‐PP‐Atto647 and/or EIPA or bafilomycin A for 3 h. Images were obtained by confocal microscopy with a 20x objective and Lysosensor Yellow/Blue dextran emission was collected in 470 ± 20, 490 ± 10 and 525 ± 25 nm band ranges. 470 ± 20 and 525 ± 25 nm were artificially colored as blue and red consecutively. 470 ± 20 (blue) and 525 ± 25 (red) emission ratios were compared with the calibration curve. 675 nm and higher rLOX‐PP‐Atto647N emission was collected and artificially colored as magenta. (Artificially magenta colored emission integral of rLOX‐PP‐Atto647N was collected to avoid overlap between Yellow/Blue dextran 10,000 and rLOX‐PP‐Atto647N emission in rLOX‐PP‐Atto647N treated samples). In rLOX‐PP‐Atto647N treated cells, the merged image on the right side shows that rLOX‐PP‐Atto647N and Lysosensor Yellow/Blue dextran 10,000 MW co‐localize. Images show endosomal pH (pH endo) measurements which change as a function of either rLOX‐PP‐Atto647N bafilomycin A1, EIPA and combinations in SCC9 cells (images were collected with EC‐Plan Neofluar 40x/1.30 oil DIC M27 objective). The excitation wavelength was 740 nm (two photon), and emission was detected in 4 channels: 470/20, 490/10 and 525/25 nm for Lysosensor Yellow/Blue dextran 10,000 MW, and 669/25 nm for rLOX‐PP‐Atto647N. In (B), SCC9 cells were treated with either both rLOX‐PP‐Atto647N and EIPA or bafilomycin A1 and EIPA to assess whether rLOX‐PP‐Atto647N or bafilomycin A1 can reverse the endosomal pH drop caused by EIPA treatment.
Figure 13
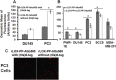
Removal of (His) 6 ‐tag increases rLOX‐PP‐Atto565 uptake, while pretreatment with rLOX‐PP decreases its uptake. In (A), DU145 cells and PC3 cells were treated with either rLOX‐PP‐Atto565 with His‐tag (gray bars) or without (His)6‐tag (white bars), and uptake levels quantified as a function of time by flow cytometry; n = 3, *, p < 0.005. (B) Cells were pre‐treated with unlabeled rLOX‐PP, followed by rLOX‐PP‐Atto56, and uptake of rLOX‐PP‐Atto565 determined; data are means ± SD, n = 3, *, two‐tailed p < 0.05 (Except in MDA‐MB‐231). (C) A representative confocal image showed that removal of (His)6‐tag increased rLOX‐PP‐Atto565 uptake in PC3 cells at 3 h (images were collected with EC‐Plan Neofluar 40x/1.30 oil DIC M27 objective).
Figure 14
Schematic representation of different intracellular uptake pathways of rLOX‐PP‐Atto565. Postulated mechanisms of rLOX‐PP‐Atto565 uptake are shown and the cell lines which use a particular uptake mechanism are indicated. They include actin‐ and PI3K‐dependent macropinocytosis, dynamin‐clathrin‐dependent endocytosis and dynamin‐caveolae‐dependent endocytosis. The effect of pH change by EIPA, bafilomycin A1 and rLOX‐PP itself on subsequent rLOX‐PP‐Atto647N trafficking was shown. *rLOX‐PP‐Atto565 uptake in DU145 cell is not PI3K‐dependent macropinocytosis.
Similar articles
- Lysyl oxidase propeptide stimulates osteoblast and osteoclast differentiation and enhances PC3 and DU145 prostate cancer cell effects on bone in vivo.
Alsulaiman M, Bais MV, Trackman PC. Alsulaiman M, et al. J Cell Commun Signal. 2016 Mar;10(1):17-31. doi: 10.1007/s12079-015-0311-9. Epub 2015 Dec 1. J Cell Commun Signal. 2016. PMID: 26627907 Free PMC article. - Effects of tumor-suppressor lysyl oxidase propeptide on prostate cancer xenograft growth and its direct interactions with DNA repair pathways.
Bais MV, Ozdener GB, Sonenshein GE, Trackman PC. Bais MV, et al. Oncogene. 2015 Apr 9;34(15):1928-37. doi: 10.1038/onc.2014.147. Epub 2014 Jun 2. Oncogene. 2015. PMID: 24882580 Free PMC article. - Characterization of recombinant lysyl oxidase propeptide.
Vora SR, Guo Y, Stephens DN, Salih E, Vu ED, Kirsch KH, Sonenshein GE, Trackman PC. Vora SR, et al. Biochemistry. 2010 Apr 6;49(13):2962-72. doi: 10.1021/bi902218p. Biochemistry. 2010. PMID: 20192271 Free PMC article. - Functions and Mechanisms of Pro-Lysyl Oxidase Processing in Cancers and Eye Pathologies with a Focus on Diabetic Retinopathy.
Trackman PC, Peymanfar Y, Roy S. Trackman PC, et al. Int J Mol Sci. 2022 May 3;23(9):5088. doi: 10.3390/ijms23095088. Int J Mol Sci. 2022. PMID: 35563478 Free PMC article. Review. - Lysyl oxidases: a novel multifunctional amine oxidase family.
Csiszar K. Csiszar K. Prog Nucleic Acid Res Mol Biol. 2001;70:1-32. doi: 10.1016/s0079-6603(01)70012-8. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11642359 Review.
Cited by
- Lysyl oxidase propeptide stimulates osteoblast and osteoclast differentiation and enhances PC3 and DU145 prostate cancer cell effects on bone in vivo.
Alsulaiman M, Bais MV, Trackman PC. Alsulaiman M, et al. J Cell Commun Signal. 2016 Mar;10(1):17-31. doi: 10.1007/s12079-015-0311-9. Epub 2015 Dec 1. J Cell Commun Signal. 2016. PMID: 26627907 Free PMC article. - Development of an AAV-Based MicroRNA Gene Therapy to Treat Machado-Joseph Disease.
Martier R, Sogorb-Gonzalez M, Stricker-Shaver J, Hübener-Schmid J, Keskin S, Klima J, Toonen LJ, Juhas S, Juhasova J, Ellederova Z, Motlik J, Haas E, van Deventer S, Konstantinova P, Nguyen HP, Evers MM. Martier R, et al. Mol Ther Methods Clin Dev. 2019 Oct 28;15:343-358. doi: 10.1016/j.omtm.2019.10.008. eCollection 2019 Dec 13. Mol Ther Methods Clin Dev. 2019. PMID: 31828177 Free PMC article. - Inhibition of angiogenesis in endothelial cells by Human Lysyl oxidase propeptide.
Nareshkumar RN, Sulochana KN, Coral K. Nareshkumar RN, et al. Sci Rep. 2018 Jul 11;8(1):10426. doi: 10.1038/s41598-018-28745-8. Sci Rep. 2018. PMID: 29993014 Free PMC article. - The challenges of oral drug delivery via nanocarriers.
Reinholz J, Landfester K, Mailänder V. Reinholz J, et al. Drug Deliv. 2018 Nov;25(1):1694-1705. doi: 10.1080/10717544.2018.1501119. Drug Deliv. 2018. PMID: 30394120 Free PMC article. Review. - Matricryptins Network with Matricellular Receptors at the Surface of Endothelial and Tumor Cells.
Ricard-Blum S, Vallet SD. Ricard-Blum S, et al. Front Pharmacol. 2016 Feb 4;7:11. doi: 10.3389/fphar.2016.00011. eCollection 2016. Front Pharmacol. 2016. PMID: 26869928 Free PMC article. Review.
References
- Ahn, S.G. , Dong, S.M. , Oshima, A. , Kim, W.H. , Lee, H.M. , Lee, S.A. , Kwon, S.H. , Lee, J.H. , Lee, J.M. , Jeong, J. , Lee, H.D. , Green, J.E. , 2013. LOXL2 expression is associated with invasiveness and negatively influences survival in breast cancer patients. Breast Cancer Res. Treat. 141, 89–99. - PMC - PubMed
- Amyere, M. , Payrastre, B. , Krause, U. , Van Der Smissen, P. , Veithen, A. , Courtoy, P.J. , 2000. Constitutive macropinocytosis in oncogene-transformed fibroblasts depends on sequential permanent activation of phosphoinositide 3-kinase and phospholipase C. Mol. Biol. Cell. 11, 3453–3467. - PMC - PubMed
- Araki, N. , Hamasaki, M. , Egami, Y. , Hatae, T. , 2006. Effect of 3-methyladenine on the fusion process of macropinosomes in EGF-stimulated A431 cells. Cell Struct. Funct. 31, 145–157. - PubMed
- Araki, N. , Hatae, T. , Furukawa, A. , Swanson, J.A. , 2003. Phosphoinositide-3-kinase-independent contractile activities associated with Fcgamma-receptor-mediated phagocytosis and macropinocytosis in macrophages. J. Cell Sci. 116, 247–257. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous