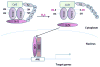H2S: A Novel Gasotransmitter that Signals by Sulfhydration - PubMed (original) (raw)
Review
H2S: A Novel Gasotransmitter that Signals by Sulfhydration
Bindu D Paul et al. Trends Biochem Sci. 2015 Nov.
Abstract
Hydrogen sulfide (H2S) is a member of the growing family of gasotransmitters. Once regarded as a noxious molecule predominantly present in the atmosphere, H2S is now known to be synthesized endogenously in mammals. H2S participates in a myriad of physiological processes ranging from regulation of blood pressure to neuroprotection. Its chemical nature precludes H2S from being stored in vesicles and acting on receptor proteins in the fashion of other chemical messengers. Thus, novel cellular mechanisms have evolved to mediate its effects. This review focuses on sulfhydration (or persulfidation), which appears to be the principal post-translational modification elicited by H2S.
Keywords: cysteine; gasotransmitter; hydrogen sulfide; sulfhydration.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
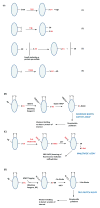
Figure 1. Potential mechanisms of protein sulfhydration and its detection.(A) Mechanisms for sulfydration/persulfidation
Sulfhydration can occur by the reaction of sulfide with oxidized cysteine residues such as cysteine sulfenic acid or disulfides (Reactions 1 and 2 respectively). Sulfhydration may also occur when an existing persulfide (on either a small molecule or a protein) reacts with a cysteine thiol (Reaction 3). Reaction of H2S2 with cysteine thiolates may also lead to sulfhydration (Reaction 4). (B) The modified biotin switch assay. The illustration depicts a protein with unmodified cysteines (-SH), sulfhydrated cysteines (-SSH) and disulfide bonded cysteines (S-S). Purified protein, or cell or tissue lysate, is incubated with methyl methanethiosulfonate (MMTS), to block unmodified cysteines. Unreacted MMTS is then removed by acetone precipitation or by gel filtration followed by treatment of the sulfhydrated protein with biotin-HPDP, which reacts with the protein at the site of sulfhydration. The biotinylated protein is enriched using streptavidin conjugates and analyzed by western blot analysis. (C) Maleimide assay. In this assay, the protein is first immunoprecipitated and treated with a fluorescent version of maleimide, which reacts with thiols under conditions that preserve the native conformation of the protein. After removing excess maleimide, the reaction mixture is treated with dithiothreitol (DTT), which reduces the disulfide bond resulting in the removal of the maleimide and a decrease in fluorescence that can be observed by SDS-PAGE. (D) The tag switch assay. The assay is a variation of the modified biotin switch assay. The reaction mixture is treated with the thiol blocking reagent (BR): methylsulfonyl benzothiazole (MSBT), followed by treatment with a methylcyanoacetate (MCA) derivative that comprises a nucleophilic component and a biotin moiety as a reporter. The biotinylated protein is then captured using streptavidin beads and analyzed by western blotting. The modifications caused by sulfide or its derivatives are shown in red.
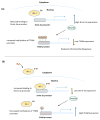
Figure 2. Sulfhydration regulates expression of genes involved in mitochondrial metabolism
(A) Interferon regulatory factor 1 (IRF-1) is a transcriptional repressor of the DNA methyltransferase 3a (Dnmt-3a). IRF-1 is regulated by sulfhydration. When H2S levels and consequently sulfhydration levels are low, IRF-1 is unable to bind its site on the Dnmt-3a promoter, leading to increased expression of Dnmt-3a, which methylates its target promoters, including the mitochondrial transcription factor A (TFAM), leading to reduced mitochondrial biogenesis. (B) When H2S production is increased, it sulfhydrates IRF-1 and enhances its interaction with the Dnmt-3a promoter to repress its expression. Consequently, methylation of the TFAM promoter is decreased, leading to a higher expression of TFAM and increased mitochondrial biogenesis.
Figure 3. Sulfhydration regulates the expression of phase II cytoprotective genes
Nuclear factor erythroid 2-related factor 2 (Nrf2) is the master regulator of a battery of genes, including the phase II genes, which respond to stressful conditions such as oxidative stress. Under basal conditions, Nrf2 is sequestered in the cytosol by kelch-like ECH-associated protein (Keap1), which targets it for proteasomal degradation involving Cul3 and E2 ubiquitin ligases. Keap1 has reactive cysteines, whose sulfhydration results in dissociation from Nrf2. Released Nrf2 translocates to the nucleus to regulate transcription of stress-responsive genes.
Figure 4. Sulfhydration in the brain
(A) Sulfhydration is dysregulated in Parkinson’s disease (PD). In normal subjects, the E3 ubiquitin ligase, Parkin, is sulfhydrated under basal conditions, which enhances its catalytic activity. Parkin mediates ubiquitylation of substrates such as α-synuclein (a component of the Lewy bodies found in PD) and targets them for degradation. In sporadic forms of PD, sulfhydration of Parkin is diminished, leading to decreased catalytic activity, which results in accumulation of toxic proteins and neurotoxicity.(B) Sulfhydration regulates synaptic function. The proinflammtory cytokine interleukin-1β (IL-1β, purple circles) plays key roles in learning and memory and is involved in promoting long term potentiation (LTP). IL-1β activates the transcription factor specificity protein 1 (SP1), which stimulates the transcription of cystathionine β-synthase (CBS), the major H2S producing enzyme in the brain, leading to sulfhydration of the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Sulfhydrated GAPDH binds to seven in absentia homolog-1 (siah1), an E3 ubiquitin ligase, which targets post-synaptic density 95 protein (PSD95) for degradation. PSD95 is a scaffolding protein that participates in synaptic functions. Degradation of PSD95 leads to spine retraction and associated cognitive deficits.
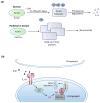
Figure 5. Reciprocity of sulfhydration and nitrosylation
Sulfhydration and nitrosylation occur on reactive cysteines and as a result frequently modify the same residue. In general, sulfhydration and nitrosylation are functionally antagonistic although there are examples where both modifications elicit the same outcome. In several instances, sulfhydration precedes nitrosylation. During inflammatory conditions (left), cystathionine γ-lyase (CSE) expression is stimulated to produce hydrogen sulfide (H2S). H2S sulfhydrates the p65 subunit of the transcription factor NF-κB and promotes its association with its coactivator, the ribosomal protein S3 (rps3), to enhance expression of cytoprotective genes. If the inflammatory signals persist, the cells produce nitric oxide (NO), which nitrosylates p65 at the same residue and inhibits its DNA binding activity and cytoprotective functions.
Similar articles
- Modes of physiologic H2S signaling in the brain and peripheral tissues.
Paul BD, Snyder SH. Paul BD, et al. Antioxid Redox Signal. 2015 Feb 10;22(5):411-23. doi: 10.1089/ars.2014.5917. Epub 2014 May 9. Antioxid Redox Signal. 2015. PMID: 24684551 Free PMC article. Review. - Protein persulfidation: Rewiring the hydrogen sulfide signaling in cell stress response.
He B, Zhang Z, Huang Z, Duan X, Wang Y, Cao J, Li L, He K, Nice EC, He W, Gao W, Shen Z. He B, et al. Biochem Pharmacol. 2023 Mar;209:115444. doi: 10.1016/j.bcp.2023.115444. Epub 2023 Feb 1. Biochem Pharmacol. 2023. PMID: 36736962 Review. - Functional and Molecular Insights of Hydrogen Sulfide Signaling and Protein Sulfhydration.
Sen N. Sen N. J Mol Biol. 2017 Feb 17;429(4):543-561. doi: 10.1016/j.jmb.2016.12.015. Epub 2016 Dec 21. J Mol Biol. 2017. PMID: 28013031 Free PMC article. Review. - Gasotransmitter hydrogen sulfide signaling in neuronal health and disease.
Paul BD, Snyder SH. Paul BD, et al. Biochem Pharmacol. 2018 Mar;149:101-109. doi: 10.1016/j.bcp.2017.11.019. Epub 2017 Dec 1. Biochem Pharmacol. 2018. PMID: 29203369 Free PMC article. Review. - Hydrogen Sulfide Biochemistry and Interplay with Other Gaseous Mediators in Mammalian Physiology.
Giuffrè A, Vicente JB. Giuffrè A, et al. Oxid Med Cell Longev. 2018 Jun 27;2018:6290931. doi: 10.1155/2018/6290931. eCollection 2018. Oxid Med Cell Longev. 2018. PMID: 30050658 Free PMC article. Review.
Cited by
- Oxidative Cysteine Post Translational Modifications Drive the Redox Code Underlying Neurodegeneration and Amyotrophic Lateral Sclerosis.
Percio A, Cicchinelli M, Masci D, Summo M, Urbani A, Greco V. Percio A, et al. Antioxidants (Basel). 2024 Jul 23;13(8):883. doi: 10.3390/antiox13080883. Antioxidants (Basel). 2024. PMID: 39199129 Free PMC article. Review. - Regulators of the transsulfuration pathway.
Sbodio JI, Snyder SH, Paul BD. Sbodio JI, et al. Br J Pharmacol. 2019 Feb;176(4):583-593. doi: 10.1111/bph.14446. Epub 2018 Aug 23. Br J Pharmacol. 2019. PMID: 30007014 Free PMC article. Review. - Sp1 S-Sulfhydration Induced by Hydrogen Sulfide Inhibits Inflammation via HDAC6/MyD88/NF-κB Signaling Pathway in Adjuvant-Induced Arthritis.
Li M, Hu W, Wang R, Li Z, Yu Y, Zhuo Y, Zhang Y, Wang Z, Qiu Y, Chen K, Ding Q, Qi W, Zhu M, Zhu Y. Li M, et al. Antioxidants (Basel). 2022 Apr 7;11(4):732. doi: 10.3390/antiox11040732. Antioxidants (Basel). 2022. PMID: 35453416 Free PMC article. - H2S mediates the vasodilator effect of endothelin-1 in the cerebral circulation.
Patel S, Fedinec AL, Liu J, Weiss MA, Pourcyrous M, Harsono M, Parfenova H, Leffler CW. Patel S, et al. Am J Physiol Heart Circ Physiol. 2018 Dec 1;315(6):H1759-H1764. doi: 10.1152/ajpheart.00451.2018. Epub 2018 Sep 28. Am J Physiol Heart Circ Physiol. 2018. PMID: 30265150 Free PMC article. - Smart H2S-Triggered/Therapeutic System (SHTS)-Based Nanomedicine.
Chen W, Ni D, Rosenkrans ZT, Cao T, Cai W. Chen W, et al. Adv Sci (Weinh). 2019 Oct 14;6(22):1901724. doi: 10.1002/advs.201901724. eCollection 2019 Nov. Adv Sci (Weinh). 2019. PMID: 31763153 Free PMC article. Review.
References
- Paul BD, Snyder SH. H2S signalling through protein sulfhydration and beyond. Nature reviews. Mol Cell Biol. 2012;13:499–507. - PubMed
- Shibuya N, et al. A novel pathway for the production of hydrogen sulfide from D-cysteine in mammalian cells. Nat Commun. 2013;4:1366. - PubMed
- Wang R. Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol Rev. 2012;92:791–896. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 MH018501/MH/NIMH NIH HHS/United States
- P50 DA000266/DA/NIDA NIH HHS/United States
- DA000266/DA/NIDA NIH HHS/United States
- MH18501/MH/NIMH NIH HHS/United States
- R01 MH018501/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources