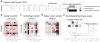Efficient genotype compression and analysis of large genetic-variation data sets - PubMed (original) (raw)
Efficient genotype compression and analysis of large genetic-variation data sets
Ryan M Layer et al. Nat Methods. 2016 Jan.
Abstract
Genotype Query Tools (GQT) is an indexing strategy that expedites analyses of genome-variation data sets in Variant Call Format based on sample genotypes, phenotypes and relationships. GQT's compressed genotype index minimizes decompression for analysis, and its performance relative to that of existing methods improves with cohort size. We show substantial (up to 443-fold) gains in performance over existing methods and demonstrate GQT's utility for exploring massive data sets involving thousands to millions of genomes. GQT can be accessed at https://github.com/ryanlayer/gqt.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Figures
Figure 1
Creation and data exploration of an individual-centric genotype index. (a) The “variant-centric” VCF standard is essentially a genotype matrix whose rows correspond to variants and columns to individuals. (b) The VCF standard is inefficient for queries across all genotypes and a subset of individuals since each variant row must be inspected (in red) to test all of the genotypes of specific individuals. (c) By transposing the matrix such that rows (data records) now represent the full set of genotypes for each individual, the data better aligns to individual-centric questions and algorithms. (d) Sorting the columns of an individual-centric matrix by alternate allele count (AC) improves compressibility. After the variants have been reorganized based on AC, all genotypes for each sample are converted to Word Aligned Hybrid compressed bitmaps (see Supplementary Note). (e) GQT will create a SQLite database of a PED file describing the familial relationships, gender, ancestry, and custom, user-defined sample descriptions. The resulting database allows GQT to quickly extract the specific compressed bitmap records in the genotype index corresponding to a query. Once the compressed bitmaps for the relevant samples are extracted, they are compared to quickly identify the subset of variant(s) that meet the genotype requirements (in this example all individuals must be heterozygous, yielding variant V10 which is denoted by an asterisk) imposed for the subset of individuals.
Figure 2
GQT query performance and applications of the genotype index. (a) Fold speedup for computing the alternate allele frequency (AF) for a targeted 10% of the 2,504 individuals in 1,000 Genomes Phase 3. The baseline was the BCFTOOLS “stats” command. Two versions of GQT output were considered, valid VCF (“GQT query”) and the count of matching variants (“GQT query -c”). (b) Speedup for finding variants having an AF of < 1% in a target 10% of individuals. The baseline was BCFTOOLS “view -C”. PLINK did not directly perform this operation and was excluded. (c) Query performance for simulated genotypes on a 100 Mb genome with between 100 and 100,000 individuals. The speedup for computing the alternate AF count for 10% of individuals is presented. (d) The speedup for finding variants having an AF of < 1% in 10% of individuals. Again, PLINK was excluded. Times reported are for 100,000 simulated genomes. Neither variant nor sample metadata were included. The metrics for 1 million individuals (est.) were estimated using a linear fit. GQT’s runtimes are similar for 2,504 individuals from the 1,000 Genomes and the simulation because the total number of genotypes is nearly identical (2,504 individuals with 84,739,846 variants and 100,000 individuals with 2,052,387 variants, respectively). (e) A principal component analysis of all variants from 1,000 Genomes Phase 3 requiring 207 minutes for 2,504 individuals, and 3 minutes for 347 AMR individuals. (f) Fst analysis of Europeans v. East Asians and Europeans v. Africans on chromosome 12.
Similar articles
- webGQT: A Shiny Server for Genotype Query Tools for Model-Based Variant Filtering.
Arumilli M, Layer RM, Hytönen MK, Lohi H. Arumilli M, et al. Front Genet. 2020 Mar 3;11:152. doi: 10.3389/fgene.2020.00152. eCollection 2020. Front Genet. 2020. PMID: 32194629 Free PMC article. - BGT: efficient and flexible genotype query across many samples.
Li H. Li H. Bioinformatics. 2016 Feb 15;32(4):590-2. doi: 10.1093/bioinformatics/btv613. Epub 2015 Oct 24. Bioinformatics. 2016. PMID: 26500154 Free PMC article. - Variant-Kudu: An Efficient Tool kit Leveraging Distributed Bitmap Index for Analysis of Massive Genetic Variation Datasets.
Fan J, Dong S, Wang B. Fan J, et al. J Comput Biol. 2020 Sep;27(9):1350-1360. doi: 10.1089/cmb.2019.0344. Epub 2020 Jan 6. J Comput Biol. 2020. PMID: 31904999 - Distributed hybrid-indexing of compressed pan-genomes for scalable and fast sequence alignment.
Maarala AI, Arasalo O, Valenzuela D, Mäkinen V, Heljanko K. Maarala AI, et al. PLoS One. 2021 Aug 3;16(8):e0255260. doi: 10.1371/journal.pone.0255260. eCollection 2021. PLoS One. 2021. PMID: 34343181 Free PMC article. - Seqminer2: an efficient tool to query and retrieve genotypes for statistical genetics analyses from biobank scale sequence dataset.
Yang L, Jiang S, Jiang B, Liu DJ, Zhan X. Yang L, et al. Bioinformatics. 2020 Dec 8;36(19):4951-4954. doi: 10.1093/bioinformatics/btaa628. Bioinformatics. 2020. PMID: 32756942 Free PMC article.
Cited by
- GSC: efficient lossless compression of VCF files with fast query.
Luo X, Chen Y, Liu L, Ding L, Li Y, Li S, Zhang Y, Zhu Z. Luo X, et al. Gigascience. 2024 Jan 2;13:giae046. doi: 10.1093/gigascience/giae046. Gigascience. 2024. PMID: 39028587 Free PMC article. - Analysis-ready VCF at Biobank scale using Zarr.
Czech E, Millar TR, White T, Jeffery B, Miles A, Tallman S, Wojdyla R, Zabad S, Hammerbacher J, Kelleher J. Czech E, et al. bioRxiv [Preprint]. 2024 Jun 12:2024.06.11.598241. doi: 10.1101/2024.06.11.598241. bioRxiv. 2024. PMID: 38915693 Free PMC article. Preprint. - Genotype Representation Graphs: Enabling Efficient Analysis of Biobank-Scale Data.
DeHaas D, Pan Z, Wei X. DeHaas D, et al. bioRxiv [Preprint]. 2024 Aug 21:2024.04.23.590800. doi: 10.1101/2024.04.23.590800. bioRxiv. 2024. PMID: 38712040 Free PMC article. Preprint. - GBC: a parallel toolkit based on highly addressable byte-encoding blocks for extremely large-scale genotypes of species.
Zhang L, Yuan Y, Peng W, Tang B, Li MJ, Gui H, Wang Q, Li M. Zhang L, et al. Genome Biol. 2023 Apr 17;24(1):76. doi: 10.1186/s13059-023-02906-z. Genome Biol. 2023. PMID: 37069653 Free PMC article. - DRAGON-Data: a platform and protocol for integrating genomic and phenotypic data across large psychiatric cohorts.
Lynham AJ, Knott S, Underwood JFG, Hubbard L, Agha SS, Bisson JI, van den Bree MBM, Chawner SJRA, Craddock N, O'Donovan M, Jones IR, Kirov G, Langley K, Martin J, Rice F, Roberts NP, Thapar A, Anney R, Owen MJ, Hall J, Pardiñas AF, Walters JTR. Lynham AJ, et al. BJPsych Open. 2023 Feb 8;9(2):e32. doi: 10.1192/bjo.2022.636. BJPsych Open. 2023. PMID: 36752340 Free PMC article.
References
- 1000 Genomes Project Consortium et al. Nature. 2012;491:56–65. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous