Chilling-Mediated DNA Methylation Changes during Dormancy and Its Release Reveal the Importance of Epigenetic Regulation during Winter Dormancy in Apple (Malus x domestica Borkh.) - PubMed (original) (raw)
Chilling-Mediated DNA Methylation Changes during Dormancy and Its Release Reveal the Importance of Epigenetic Regulation during Winter Dormancy in Apple (Malus x domestica Borkh.)
Gulshan Kumar et al. PLoS One. 2016.
Abstract
Winter dormancy is a well known mechanism adopted by temperate plants, to mitigate the chilling temperature of winters. However, acquisition of sufficient chilling during winter dormancy ensures the normal phenological traits in subsequent growing period. Thus, low temperature appears to play crucial roles in growth and development of temperate plants. Apple, being an important temperate fruit crop, also requires sufficient chilling to release winter dormancy and normal phenological traits, which are often associated with yield and quality of fruits. DNA cytosine methylation is one of the important epigenetic modifications which remarkably affect the gene expression during various developmental and adaptive processes. In present study, methylation sensitive amplified polymorphism was employed to assess the changes in cytosine methylation during dormancy, active growth and fruit set in apple, under differential chilling conditions. Under high chill conditions, total methylation was decreased from 27.2% in dormant bud to 21.0% in fruit set stage, while no significant reduction was found under low chill conditions. Moreover, the demethylation was found to be decreased, while methylation increased from dormant bud to fruit set stage under low chill as compared to high chill conditions. In addition, RNA-Seq analysis showed high expression of DNA methyltransferases and histone methyltransferases during dormancy and fruit set, and low expression of DNA glcosylases during active growth under low chill conditions, which was in accordance with changes in methylation patterns. The RNA-Seq data of 47 genes associated with MSAP fragments involved in cellular metabolism, stress response, antioxidant system and transcriptional regulation showed correlation between methylation and their expression. Similarly, bisulfite sequencing and qRT-PCR analysis of selected genes also showed correlation between gene body methylation and gene expression. Moreover, significant association between chilling and methylation changes was observed, which suggested that chilling acquisition during dormancy in apple is likely to affect the epigenetic regulation through DNA methylation.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Fig 1. The graph showing the average of minimum and maximum daily temperature during November to April for three consecutive years (2011–2014) at both the sampling locations.
The HC and LC represent the high chill condition (Palchan) and low chill condition (Seobag), respectively. The min. and max. represent the minimum and maximum temperature, respectively.
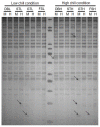
Fig 2. A representative gel image of MSAP assay showing polymorphic fragments in different samples.
lane H: DNA digested with _Eco_RI–_Hpa_II; lane M: DNA digested with _Eco_RI–_Msp_I. The four developmental stages viz. Dormant bud (DB), Silver tip (ST), Green tip (GT) and Initial fruit set (FS) in apple, under low (L) chill and high (H) chill conditions, were used for MSAP analysis. The arrow marks show presence of polymorphic fragments.
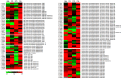
Fig 3. Heat map representation of relative expression based on FPKM values of differentially methylated fragments in various developmental tissues as identified in MSAP analysis.
The majority of demethylated fragments showed upregulation under low chill conditions as compared to high chill conditions. The relative expression of majority of methylated fragments was downregulated. The relative FPKM values were used to generate the heat-map using MeV4. The color scale at the bottom represents the expression level, where red, green and black colors indicate upregulation, downregulation and unaltered expression, respectively. Contig number (starting with C_) and their annotation are given on the left and right side of the heat map, respectively.
Fig 4. Heat map representation of relative expression based on FPKM values of DNA methyltransferases, DNA glycosylases and histone methyltransferases in various developmental tissues.
The relative FPKM values were used to generate the heat-map using MeV4. The color scale at the bottom represents the expression level, where red, green and black colors indicate upregulation, downregulation and unaltered expression, respectively. Contig number (starting with C_) and their annotation are given on the left and right side of the heat map, respectively.
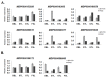
Fig 5
Relative expression of randomly selected (A) demethylated and (B) methylated genes analyzed through qRT-PCR. Pearson correlation coefficient between fold change of FPKM based expression and qRT-PCR was 0.431 (p-value<0.0005).
Similar articles
- Chilling Affects Phytohormone and Post-Embryonic Development Pathways during Bud Break and Fruit Set in Apple (Malus domestica Borkh.).
Kumar G, Gupta K, Pathania S, Swarnkar MK, Rattan UK, Singh G, Sharma RK, Singh AK. Kumar G, et al. Sci Rep. 2017 Feb 15;7:42593. doi: 10.1038/srep42593. Sci Rep. 2017. PMID: 28198417 Free PMC article. - Potential vulnerability of Moroccan apple orchard to climate change-induced phenological perturbations: effects on yields and fruit quality.
El Yaacoubi A, El Jaouhari N, Bourioug M, El Youssfi L, Cherroud S, Bouabid R, Chaoui M, Abouabdillah A. El Yaacoubi A, et al. Int J Biometeorol. 2020 Mar;64(3):377-387. doi: 10.1007/s00484-019-01821-y. Epub 2019 Nov 26. Int J Biometeorol. 2020. PMID: 31773321 - Comparative RNA-Sequencing and DNA Methylation Analyses of Apple (Malus domestica Borkh.) Buds with Diverse Flowering Capabilities Reveal Novel Insights into the Regulatory Mechanisms of Flower Bud Formation.
Xing L, Li Y, Qi S, Zhang C, Ma W, Zuo X, Liang J, Gao C, Jia P, Shah K, Zhang D, An N, Zhao C, Han M, Zhao J. Xing L, et al. Plant Cell Physiol. 2019 Aug 1;60(8):1702-1721. doi: 10.1093/pcp/pcz080. Plant Cell Physiol. 2019. PMID: 31077318 - Insights into flowering mechanisms in apple (Malus × domestica Borkh.) amidst climate change: An exploration of genetic and epigenetic factors.
Kumar A, Mushtaq M, Kumar P, Sharma DP, Gahlaut V. Kumar A, et al. Biochim Biophys Acta Gen Subj. 2024 May;1868(5):130593. doi: 10.1016/j.bbagen.2024.130593. Epub 2024 Feb 24. Biochim Biophys Acta Gen Subj. 2024. PMID: 38408683 Review. - Genetic and molecular regulation of chilling requirements in pear: breeding for climate change resilience.
Gabay G, Flaishman MA. Gabay G, et al. Front Plant Sci. 2024 Apr 26;15:1347527. doi: 10.3389/fpls.2024.1347527. eCollection 2024. Front Plant Sci. 2024. PMID: 38736438 Free PMC article. Review.
Cited by
- Molecular Mechanisms Underlying Freezing Tolerance in Plants: Implications for Cryopreservation.
Białoskórska M, Rucińska A, Boczkowska M. Białoskórska M, et al. Int J Mol Sci. 2024 Sep 20;25(18):10110. doi: 10.3390/ijms251810110. Int J Mol Sci. 2024. PMID: 39337593 Free PMC article. Review. - Unravelling the Epigenetic Code: DNA Methylation in Plants and Its Role in Stress Response.
Talarico E, Zambelli A, Araniti F, Greco E, Chiappetta A, Bruno L. Talarico E, et al. Epigenomes. 2024 Aug 8;8(3):30. doi: 10.3390/epigenomes8030030. Epigenomes. 2024. PMID: 39189256 Free PMC article. Review. - Molecular Mechanisms of Seasonal Gene Expression in Trees.
Chu X, Wang M, Fan Z, Li J, Yin H. Chu X, et al. Int J Mol Sci. 2024 Jan 30;25(3):1666. doi: 10.3390/ijms25031666. Int J Mol Sci. 2024. PMID: 38338945 Free PMC article. Review. - Mechanisms of Plant Epigenetic Regulation in Response to Plant Stress: Recent Discoveries and Implications.
Abdulraheem MI, Xiong Y, Moshood AY, Cadenas-Pliego G, Zhang H, Hu J. Abdulraheem MI, et al. Plants (Basel). 2024 Jan 7;13(2):163. doi: 10.3390/plants13020163. Plants (Basel). 2024. PMID: 38256717 Free PMC article. Review. - DNA cytosine methylation dynamics and functional roles in horticultural crops.
Liu P, Liu R, Xu Y, Zhang C, Niu Q, Lang Z. Liu P, et al. Hortic Res. 2023 Aug 29;10(10):uhad170. doi: 10.1093/hr/uhad170. eCollection 2023 Oct. Hortic Res. 2023. PMID: 38025976 Free PMC article.
References
- Bond DM, Finnegan EJ. Passing the message on: inheritance of epigenetic traits. Trends Plant Sci. 2007; 12:211–216. - PubMed
- Causevic A, Delaunay A, Ounnar S, Righezza M, Delmotte F, Brignolas F, et al. DNA methylating and demethylating treatments modify phenotype and cell wall differentiation state in sugarbeet cell lines. Plant Physiol Biochem. 2005; 43:681–691. - PubMed
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007; 447:418–424. - PubMed
Publication types
MeSH terms
Grants and funding
This study was supported by the grants from Council of Scientific and Industrial Research, New Delhi, in the form of Network Project PlaGen (BSC0107) to AKS at CSIR-IHBT, Palampur.
LinkOut - more resources
Full Text Sources
Other Literature Sources