Faustoviruses: Comparative Genomics of New Megavirales Family Members - PubMed (original) (raw)
Faustoviruses: Comparative Genomics of New Megavirales Family Members
Samia Benamar et al. Front Microbiol. 2016.
Abstract
An emerging interest for the giant virus discovery process, genome sequencing and analysis has allowed an expansion of the number of known Megavirales members. Using the protist Vermamoeba sp. as cell support, a new giant virus named Faustovirus has been isolated. In this study, we describe the genome sequences of nine Faustoviruses and build a genomic comparison in order to have a comprehensive overview of genomic composition and diversity among this new virus family. The average sequence length of these viruses is 467,592.44 bp (ranging from 455,803 to 491,024 bp), making them the fourth largest Megavirales genome after Mimiviruses, Pandoraviruses, and Pithovirus sibericum. Faustovirus genomes displayed an average G+C content of 37.14 % (ranging from 36.22 to 39.59%) which is close to the G+C content range of the Asfarviridae genomes (38%). The proportion of best matches and the phylogenetic analysis suggest a shared origin with Asfarviridae without belonging to the same family. The core-gene-based phylogeny of Faustoviruses study has identified four lineages. These results were confirmed by the analysis of amino acids and COGs category distribution. The diversity of the gene composition of these lineages is mainly explained by gene deletion or acquisition and some exceptions for gene duplications. The high proportion of best matches from Bacteria and Phycodnaviridae on the pan-genome and unique genes may be explained by an interaction occurring after the separation of the lineages. The Faustovirus core-genome appears to consolidate the surrounding of 207 genes whereas the pan-genome is described as an open pan-genome, its enrichment via the discovery of new Faustoviruses is required to better seize all the genomic diversity of this family.
Keywords: Faustovirus; genomic comparison; giant viruses; pan-genome; phylogenetic.
Figures
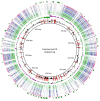
FIGURE 1
Regions of variability among the Faustovirus genomes. BLAST based genome showing regions of variability of Faustovirus E9 and the other Faustoviruses. From outer to inner circle: Unique genes of Faustovirus E9, Faustovirus Liban, Faustovirus D5a, Faustovirus E24, Faustovirus E23, Faustovirus E12, Faustovirus D5b, Faustovirus D6, Faustovirus D3, GC Skew and GC content. The unique genes of Faustovirus E9 were shown in the first circle (Green) and superimposed (in red) on the GC content circle.
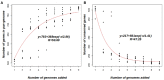
FIGURE 2
Faustovirus pan-genome and core-genome. (A) Accumulation curve for the total number of genes plotted against the number of genomes analyzed. (B) Accumulation curves for the number of genes in common plotted against the number of added genomes, the coregenes count 148 hepothetical protein (71.5%). The deduced mathematical function and the residual standard error are also reported.
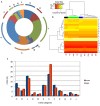
FIGURE 3
Comparative genomics of fully sequenced Faustovirus genomes and core-gene-based phylogeny. (A) Protein clusters and viruses’ connection, nodes correspond to protein clusters (small nodes) or viruses. Edges signify presence of a protein cluster in the virus. Each protein cluster is connected to the virus having a protein therein. Viruses and clusters are placed depending on the links between them. Coregenes are in the middle of the figure and the single genes on the suburbs. (B) Venn diagram representing the orthologous and unique gene families of the four lineages. (C) Phylogenetic tree based on the 207 core-genes. Maximum likelihood based on the phylogenetic analysis of a concatenated set of 207 genes.
FIGURE 4
Phylogenetic reconstruction based on concatenated A32-like packaging ATPase and DNA polymerase in Megavirales members. Protein sequences were aligned using the MUSCLE program with default parameters; all the alignments were conserved, even the columns containing a large fraction of gaps. The maximum-likelihood phylogenetic reconstruction was performed using FastTree. The Faustoviruses cluster with members of Asfarviridae and Poxviridae. The tree is unrooted. Each Megavirales family was represented by a specific color. The values near the branches (green) are bootstrap values.
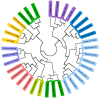
FIGURE 5
Analysis of amino acids and COGs category distribution. (A) Cluster of Orthologs (COG) classification of the families of orthologs. The most abundant families have also been indicated and they are assigned to housekeeping functions. From outer to inner circle: Faustovirus D3, Faustovirus D6, Faustovirus D5b, Faustovirus E12, Faustovirus E24, Faustovirus E23, Faustovirus D5a, Faustovirus Liban and Faustovirus E9. (B) Hierarchical clustering heat map representing the variability of Faustoviruses in terms of amino acid composition for nine complete Faustovirus genomes. (C) The COG category comparison between the core and the pan-genome of Faustoviruses.
Similar articles
- Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae.
Reteno DG, Benamar S, Khalil JB, Andreani J, Armstrong N, Klose T, Rossmann M, Colson P, Raoult D, La Scola B. Reteno DG, et al. J Virol. 2015 Jul;89(13):6585-94. doi: 10.1128/JVI.00115-15. J Virol. 2015. PMID: 25878099 Free PMC article. - Virus Genomes from Deep Sea Sediments Expand the Ocean Megavirome and Support Independent Origins of Viral Gigantism.
Bäckström D, Yutin N, Jørgensen SL, Dharamshi J, Homa F, Zaremba-Niedwiedzka K, Spang A, Wolf YI, Koonin EV, Ettema TJG. Bäckström D, et al. mBio. 2019 Mar 5;10(2):e02497-18. doi: 10.1128/mBio.02497-18. mBio. 2019. PMID: 30837339 Free PMC article. - Pacmanvirus, a New Giant Icosahedral Virus at the Crossroads between Asfarviridae and Faustoviruses.
Andreani J, Khalil JYB, Sevvana M, Benamar S, Di Pinto F, Bitam I, Colson P, Klose T, Rossmann MG, Raoult D, La Scola B. Andreani J, et al. J Virol. 2017 Jun 26;91(14):e00212-17. doi: 10.1128/JVI.00212-17. Print 2017 Jul 15. J Virol. 2017. PMID: 28446673 Free PMC article. - Giant Viruses of Amoebas: An Update.
Aherfi S, Colson P, La Scola B, Raoult D. Aherfi S, et al. Front Microbiol. 2016 Mar 22;7:349. doi: 10.3389/fmicb.2016.00349. eCollection 2016. Front Microbiol. 2016. PMID: 27047465 Free PMC article. Review. - Evolution of the Large Nucleocytoplasmic DNA Viruses of Eukaryotes and Convergent Origins of Viral Gigantism.
Koonin EV, Yutin N. Koonin EV, et al. Adv Virus Res. 2019;103:167-202. doi: 10.1016/bs.aivir.2018.09.002. Epub 2018 Nov 10. Adv Virus Res. 2019. PMID: 30635076 Review.
Cited by
- Ancestrality and Mosaicism of Giant Viruses Supporting the Definition of the Fourth TRUC of Microbes.
Colson P, Levasseur A, La Scola B, Sharma V, Nasir A, Pontarotti P, Caetano-Anollés G, Raoult D. Colson P, et al. Front Microbiol. 2018 Nov 27;9:2668. doi: 10.3389/fmicb.2018.02668. eCollection 2018. Front Microbiol. 2018. PMID: 30538677 Free PMC article. - Diversity of Giant Viruses Infecting Vermamoeba vermiformis.
Geballa-Koukoulas K, La Scola B, Blanc G, Andreani J. Geballa-Koukoulas K, et al. Front Microbiol. 2022 Apr 22;13:808499. doi: 10.3389/fmicb.2022.808499. eCollection 2022. Front Microbiol. 2022. PMID: 35602053 Free PMC article. Review. - Orpheovirus IHUMI-LCC2: A New Virus among the Giant Viruses.
Andreani J, Khalil JYB, Baptiste E, Hasni I, Michelle C, Raoult D, Levasseur A, La Scola B. Andreani J, et al. Front Microbiol. 2018 Jan 22;8:2643. doi: 10.3389/fmicb.2017.02643. eCollection 2017. Front Microbiol. 2018. PMID: 29403444 Free PMC article. - Genomic and metagenomic signatures of giant viruses are ubiquitous in water samples from sewage, inland lake, waste water treatment plant, and municipal water supply in Mumbai, India.
Chatterjee A, Sicheritz-Pontén T, Yadav R, Kondabagil K. Chatterjee A, et al. Sci Rep. 2019 Mar 6;9(1):3690. doi: 10.1038/s41598-019-40171-y. Sci Rep. 2019. PMID: 30842490 Free PMC article. - Promoter Motifs in NCLDVs: An Evolutionary Perspective.
Oliveira GP, Andrade AC, Rodrigues RA, Arantes TS, Boratto PV, Silva LK, Dornas FP, Trindade GS, Drumond BP, La Scola B, Kroon EG, Abrahão JS. Oliveira GP, et al. Viruses. 2017 Jan 20;9(1):16. doi: 10.3390/v9010016. Viruses. 2017. PMID: 28117683 Free PMC article. Review.
References
- Boetzer M., Henkel C. V., Jansen H. J., Butler D., Pirovano W. (2011). Scaffolding pre-assembled contigs using SSPACE. J. Gerontol. 27 578–579. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources