Role of TET enzymes in DNA methylation, development, and cancer - PubMed (original) (raw)
Review
Role of TET enzymes in DNA methylation, development, and cancer
Kasper Dindler Rasmussen et al. Genes Dev. 2016.
Abstract
The pattern of DNA methylation at cytosine bases in the genome is tightly linked to gene expression, and DNA methylation abnormalities are often observed in diseases. The ten eleven translocation (TET) enzymes oxidize 5-methylcytosines (5mCs) and promote locus-specific reversal of DNA methylation. TET genes, and especially TET2, are frequently mutated in various cancers, but how the TET proteins contribute to prevent the onset and maintenance of these malignancies is largely unknown. Here, we highlight recent advances in understanding the physiological function of the TET proteins and their role in regulating DNA methylation and transcription. In addition, we discuss some of the key outstanding questions in the field.
Keywords: DNA demethylation; DNA methylation; TET; hydroxymethyl cytosine; hematopoiesis; leukemia.
© 2016 Rasmussen and Helin; Published by Cold Spring Harbor Laboratory Press.
Figures
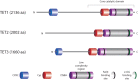
Figure 1.
Domain structure of TET proteins. The C-terminal core catalytic domain shared by all TET enzymes consists of the DSBH domain, a cysteine-rich (Cys) domain, and binding sites for the Fe(II) and 2-OG cofactors. The DSBH domain contains a large low-complexity region of unknown function. TET1 and TET3 have an N-terminal CXXC domain that can bind directly to DNA and facilitate recruitment to genomic target sites.
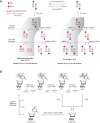
Figure 2.
Pathways of DNA demethylation mediated by TET enzymes. (A) Model of passive replication-dependent DNA demethylation. The diagrams illustrate the two opposite fates of a hemihydroxymethylated CpG dinucleotide through two rounds of DNA replication. The left panel shows the replication-dependent DNA demethylation, in which DNA methylation is lost on the DNA strand opposite to 5hmC. The right panel shows the reverse outcome, in which maintenance of DNA methylation is achieved by DNMT1 in complex with UHRF1/2 or DNMT3A/B. The relative extent of these two opposing outcomes of 5hmC deposition in vivo remains to be determined and is likely influenced by global as well as locus-specific factors. In both cases, newly replicated DNA dilutes 5hmC during cell division. (B) Model of active DNA demethylation by a TET/TDG (thymine–DNA–glycosylase)/BER (base excision repair)-dependent pathway. A cytosine base can be methylated by the DNA methylation machinery (DNMT1 or DNMT3A/B) to form 5mC, which in turn can be iteratively oxidized by TET enzymes to produce 5hmC, 5fC, and 5caC. TDG then recognizes 5fC and 5caC, and the oxidized cytosine base is excised. This yields an abasic site that is repaired by BER and results in restoration of the unmodified cytosine state. Additional pathways of active DNA demethylation have been suggested (for review, see Wu and Zhang 2014). (SAH) _S_-adenosyl-homocysteine; (SAM) _S_-adenosyl-methionine.
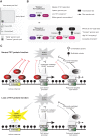
Figure 3.
The dynamics of TET-mediated cytosine oxidation and its impact on DNA methylation patterns on regulatory elements in vivo. (A) Diagram illustrating the relative abundance of cytosines in the genome with “stable” levels of modifications as well as the small “transient” fractions that undergo rapid turnover and removal by active DNA demethylation processes. The stable levels of genomic 5caC have not been experimentally determined, as they are often under the detection limit. (B) Model illustrating the dual role of TET enzymes. The rate constant of TET enzymes to produce 5hmC from 5mC is fast, whereas the rate constants for further oxidation to 5fC and 5caC are considerably slower. On a majority of TET target sites, 5mC is oxidized and persists as “stable” 5hmC, likely due to low TET residence time (“stalling TET modification”). On a minority of sites, preferably located in open chromatin associated with regulatory elements, 5mC is oxidized iteratively to 5fC and 5caC and results in net DNA demethylation by the TET/TDG/BER pathway (“processive TET modification”). (C) Model illustrating the impact of loss of TET protein function on different regulatory elements. The protection of active and inactive CpG island promoters by the TET proteins is only one of several mechanisms by which they are protected against ectopic DNA methylation. In contrast, distal regulatory elements such as active enhancers are vulnerable to aberrant DNMT activity and become DNA hypermethylated upon loss of TET activity. (PcG) Polycomb group.
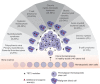
Figure 4.
Illustration of the mutational landscape of TET2 in hematological diseases. A somatic mutation in TET2 results in premalignant hematopoiesis and clonal expansion. Additional oncogenic events cooperate with the initial TET2 mutation to drive the onset of a wide variety of hematopoietic malignancies. The frequencies of TET2 mutations in the different patient groups are indicated (for review, see Scourzic et al. 2015).
Similar articles
- Functions of TET Proteins in Hematopoietic Transformation.
Han JA, An J, Ko M. Han JA, et al. Mol Cells. 2015 Nov;38(11):925-35. doi: 10.14348/molcells.2015.0294. Epub 2015 Nov 10. Mol Cells. 2015. PMID: 26552488 Free PMC article. Review. - TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling.
Dai HQ, Wang BA, Yang L, Chen JJ, Zhu GC, Sun ML, Ge H, Wang R, Chapman DL, Tang F, Sun X, Xu GL. Dai HQ, et al. Nature. 2016 Oct 27;538(7626):528-532. doi: 10.1038/nature20095. Epub 2016 Oct 19. Nature. 2016. PMID: 27760115 - TET proteins and 5-methylcytosine oxidation in hematological cancers.
Ko M, An J, Pastor WA, Koralov SB, Rajewsky K, Rao A. Ko M, et al. Immunol Rev. 2015 Jan;263(1):6-21. doi: 10.1111/imr.12239. Immunol Rev. 2015. PMID: 25510268 Free PMC article. Review. - Development of a rapid mass spectrometric method for the analysis of ten-eleven translocation enzymes.
Graves C, Islam K. Graves C, et al. Methods Enzymol. 2024;703:87-120. doi: 10.1016/bs.mie.2024.06.001. Epub 2024 Jun 20. Methods Enzymol. 2024. PMID: 39261005 - TET Enzymes in the Immune System: From DNA Demethylation to Immunotherapy, Inflammation, and Cancer.
López-Moyado IF, Ko M, Hogan PG, Rao A. López-Moyado IF, et al. Annu Rev Immunol. 2024 Jun;42(1):455-488. doi: 10.1146/annurev-immunol-080223-044610. Epub 2024 Jun 14. Annu Rev Immunol. 2024. PMID: 38360546 Review.
Cited by
- Base-excision repair pathway shapes 5-methylcytosine deamination signatures in pan-cancer genomes.
Silveira AB, Houy A, Ganier O, Özemek B, Vanhuele S, Vincent-Salomon A, Cassoux N, Mariani P, Pierron G, Leyvraz S, Rieke D, Picca A, Bielle F, Yaspo ML, Rodrigues M, Stern MH. Silveira AB, et al. Nat Commun. 2024 Nov 14;15(1):9864. doi: 10.1038/s41467-024-54223-z. Nat Commun. 2024. PMID: 39543136 Free PMC article. - TET2 regulates early and late transitions in exhausted CD8+ T cell differentiation and limits CAR T cell function.
Dimitri AJ, Baxter AE, Chen GM, Hopkins CR, Rouin GT, Huang H, Kong W, Holliday CH, Wiebking V, Bartoszek R, Drury S, Dalton K, Koucky OM, Chen Z, Giles JR, Dils AT, Jung IY, O'Connor R, Collins S, Everett JK, Amses K, Sherrill-Mix S, Chandra A, Goldman N, Vahedi G, Jadlowsky JK, Young RM, Melenhorst JJ, Maude SL, Levine BL, Frey NV, Berger SL, Grupp SA, Porter DL, Herbst F, Porteus MH, Carty SA, Bushman FD, Weber EW, Wherry EJ, Jordan MS, Fraietta JA. Dimitri AJ, et al. Sci Adv. 2024 Nov 15;10(46):eadp9371. doi: 10.1126/sciadv.adp9371. Epub 2024 Nov 13. Sci Adv. 2024. PMID: 39536093 Free PMC article. - TET2 germline variants promote kidney disease by impairing DNA repair and activating cytosolic nucleotide sensors.
Liang X, Liu H, Hu H, Ha E, Zhou J, Abedini A, Sanchez-Navarro A, Klötzer KA, Susztak K. Liang X, et al. Nat Commun. 2024 Nov 7;15(1):9621. doi: 10.1038/s41467-024-53798-x. Nat Commun. 2024. PMID: 39511169 Free PMC article. - Hypermethylation of CDKN2A CpG island drives resistance to PRC2 inhibitors in SWI/SNF loss-of-function tumors.
Wang X, Wang Y, Xie M, Ma S, Zhang Y, Wang L, Ge Y, Li G, Zhao M, Chen S, Yan C, Zhang H, Sun W. Wang X, et al. Cell Death Dis. 2024 Nov 5;15(11):794. doi: 10.1038/s41419-024-07109-3. Cell Death Dis. 2024. PMID: 39500892 Free PMC article. - Epigenetic Regulation in Myocardial Fibroblasts and Its Impact on Cardiovascular Diseases.
Komal S, Gao Y, Wang ZM, Yu QW, Wang P, Zhang LR, Han SN. Komal S, et al. Pharmaceuticals (Basel). 2024 Oct 10;17(10):1353. doi: 10.3390/ph17101353. Pharmaceuticals (Basel). 2024. PMID: 39458994 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources