Bayesian methods outperform parsimony but at the expense of precision in the estimation of phylogeny from discrete morphological data - PubMed (original) (raw)
Bayesian methods outperform parsimony but at the expense of precision in the estimation of phylogeny from discrete morphological data
Joseph E O'Reilly et al. Biol Lett. 2016 Apr.
Abstract
Different analytical methods can yield competing interpretations of evolutionary history and, currently, there is no definitive method for phylogenetic reconstruction using morphological data. Parsimony has been the primary method for analysing morphological data, but there has been a resurgence of interest in the likelihood-based Mk-model. Here, we test the performance of the Bayesian implementation of the Mk-model relative to both equal and implied-weight implementations of parsimony. Using simulated morphological data, we demonstrate that the Mk-model outperforms equal-weights parsimony in terms of topological accuracy, and implied-weights performs the most poorly. However, the Mk-model produces phylogenies that have less resolution than parsimony methods. This difference in the accuracy and precision of parsimony and Bayesian approaches to topology estimation needs to be considered when selecting a method for phylogeny reconstruction.
Keywords: Bayesian; likelihood; morphology; parsimony; phylogenetics.
© 2016 The Authors.
Figures
Figure 1.
Mk tree reconstructions (blue) outperform equal-weights parsimony (grey) and implied-weights parsimony (green) for 100, 350 and 1000 characters (a,c,e,g), and these differences remain in the subset of the simulated data matrices that exhibit realistic levels of homoplasy (b,d,f,h). Bars above the plots mark the 95th percentile range for each method, and dashed vertical lines show the median values. Percentage topology error (g,h) is the Robinson–Foulds value of the reconstructed tree compared with the worst possible value, as shown in [5].
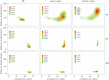
Figure 2.
The Mk model exhibits higher accuracy with lower precision than parsimony methods; these results are less clear as more characters are added. Contour plots of Robinson–Foulds distances against the number of resolved nodes in each tree; the contours represent the density of the distribution of trees.
Similar articles
- Uncertain-tree: discriminating among competing approaches to the phylogenetic analysis of phenotype data.
Puttick MN, O'Reilly JE, Tanner AR, Fleming JF, Clark J, Holloway L, Lozano-Fernandez J, Parry LA, Tarver JE, Pisani D, Donoghue PC. Puttick MN, et al. Proc Biol Sci. 2017 Jan 11;284(1846):20162290. doi: 10.1098/rspb.2016.2290. Proc Biol Sci. 2017. PMID: 28077778 Free PMC article. - Bayesian analysis using a simple likelihood model outperforms parsimony for estimation of phylogeny from discrete morphological data.
Wright AM, Hillis DM. Wright AM, et al. PLoS One. 2014 Oct 3;9(10):e109210. doi: 10.1371/journal.pone.0109210. eCollection 2014. PLoS One. 2014. PMID: 25279853 Free PMC article. - Parsimony, not Bayesian analysis, recovers more stratigraphically congruent phylogenetic trees.
Sansom RS, Choate PG, Keating JN, Randle E. Sansom RS, et al. Biol Lett. 2018 Jun;14(6):20180263. doi: 10.1098/rsbl.2018.0263. Biol Lett. 2018. PMID: 29925561 Free PMC article. - Comparative evaluation of maximum parsimony and Bayesian phylogenetic reconstruction using empirical morphological data.
Schrago CG, Aguiar BO, Mello B. Schrago CG, et al. J Evol Biol. 2018 Oct;31(10):1477-1484. doi: 10.1111/jeb.13344. Epub 2018 Jul 18. J Evol Biol. 2018. PMID: 29957887 - Bayesian and parsimony approaches reconstruct informative trees from simulated morphological datasets.
Smith MR. Smith MR. Biol Lett. 2019 Feb 28;15(2):20180632. doi: 10.1098/rsbl.2018.0632. Biol Lett. 2019. PMID: 30958126 Free PMC article.
Cited by
- The first iguanian lizard from the Mesozoic of Africa.
Apesteguía S, Daza JD, Simões TR, Rage JC. Apesteguía S, et al. R Soc Open Sci. 2016 Sep 21;3(9):160462. doi: 10.1098/rsos.160462. eCollection 2016 Sep. R Soc Open Sci. 2016. PMID: 27703708 Free PMC article. - Multiple optimality criteria support Ornithoscelida.
Parry LA, Baron MG, Vinther J. Parry LA, et al. R Soc Open Sci. 2017 Oct 25;4(10):170833. doi: 10.1098/rsos.170833. eCollection 2017 Oct. R Soc Open Sci. 2017. PMID: 29134086 Free PMC article. - Uncertain-tree: discriminating among competing approaches to the phylogenetic analysis of phenotype data.
Puttick MN, O'Reilly JE, Tanner AR, Fleming JF, Clark J, Holloway L, Lozano-Fernandez J, Parry LA, Tarver JE, Pisani D, Donoghue PC. Puttick MN, et al. Proc Biol Sci. 2017 Jan 11;284(1846):20162290. doi: 10.1098/rspb.2016.2290. Proc Biol Sci. 2017. PMID: 28077778 Free PMC article. - Fossils improve phylogenetic analyses of morphological characters.
Mongiardino Koch N, Garwood RJ, Parry LA. Mongiardino Koch N, et al. Proc Biol Sci. 2021 May 12;288(1950):20210044. doi: 10.1098/rspb.2021.0044. Epub 2021 May 5. Proc Biol Sci. 2021. PMID: 33947239 Free PMC article. - The unbearable uncertainty of panarthropod relationships.
Wu R, Pisani D, Donoghue PCJ. Wu R, et al. Biol Lett. 2023 Jan;19(1):20220497. doi: 10.1098/rsbl.2022.0497. Epub 2023 Jan 11. Biol Lett. 2023. PMID: 36628953 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- BB/J00538X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/N000919/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources